Entry Version:
Citation:
Pancreapedia: Exocrine Pancreas Knowledge Base, DOI: 10.3998/panc.2014.6
| Attachment | Size |
|---|---|
| 716.02 KB |
Gene symbols: RYR1, RYR2, RYR3
1. General Information
1.1 Size and Evolution
The ryanodine receptor (RyR) is a selective Ca2+ release channel that is localized to the endoplasmic reticulum (ER) in both excitable and non-excitable cells. The RyR owes its name to the plant alkaloid and high affinity ligand ryanodine. The functional RyR exists as a tetramer composed of a single unit of approximately 565kDa. To date, it is the largest known ion channel. In mammals, there are three RyR isoforms that share approximately 70% homology and several biophysical properties (26). RyR1 is the dominant Ca2+ release channel present on sarcoplasmic reticulum (SR) in skeletal muscles (104, 127), RyR2 is predominant in cardiac muscle (73), and in brain there is more general expression of all three isoforms (29, 36, 67, 73, 126).
Based on phylogenetic analysis, it is likely that all three RyR isoforms evolved from a single ancestor. In addition, there is evidence to suggest that the RyR as well as another intracellular Ca2+ channel the inositol 1,4,5-trisphosphate receptor (IP3R) evolved from a common ancestral gene (107). Sequence analysis reveals a high degree of similarity in the channel pore region (107), supporting the common ancestor theory. This structural homology is also present between the pore-forming region of RyRs and other ion channels, which include voltage gated ion channels, cyclic nucleotide gated channels, and the transient receptor potential channels (TRP) (41). Phylogenetic studies also reveal that certain RyR domains are highly conserved from worms to humans. For instance, there is 35-40% homology at the amino acid level between C. elegans and vertebrates, and the functional domains including the leucine/isoleucine zipper, the pore region, and the transmembrane domains are highly conserved in all three RyR isoforms. Mutations in these domains alter channel functions such as caffeine-induced Ca2+ release, ryanodine binding, single channel conductance, and cation selectivity (31, 122).
1.2 The RyR genes
The organization of the phylogenetic tree supports the model than an expansion of the vertebrate gene family was associated with an initial duplication of the RyR1 gene, since only one RyR gene is found in invertebrates. Non-mammalian vertebrates express two RyR genes (80). Further, it appears that a second duplication occurred in mammals, which distinguishes them from other vertebrates. In mammals, three RyR isoforms are transcribed from three separate genes. In humans the gene for RYR1 is located on chromosome 19q13.2 and spans 104 exons. The RYR2 gene is located on chromosome 1q43 and spans 102 exons, and the RYR3 gene is located on chromosome 15q13.3-14 and spans 103 exons.
An additional means of generating diversity in RyR channels is through alternative splicing (30, 73, 106, 107, 114). Although there are reports that splice variants, for example, of RyR3 exhibit reduced caffeine sensitivity (49), relatively little is known about the functional properties of alternatively spliced RyR channels.
1.3 Three-dimensional structure and ligand binding
Three-dimensional (3D) structural information is essential to an understanding of the functional properties of RyRs. While there are no published full length crystal structures of the RyR, there are several crystal structures of the N-terminal domain, and they reveal structural similarities with the IP3R (62, 92). Cryo-electron microscopy (EM) generated 3D reconstructions of RyR reveal a highly conserved structure, having the overall shape of a mushroom and consisting of two major components: a large, square prism-shaped cytoplasmic assembly composed of at least 10 distinct domains, and a differentiated small transmembrane assembly (Figure 1). Sequence analysis (104, 106) and biochemical studies (20, 37) demonstrate that the N-terminus (~4000-4500 amino acid residues) contains the large cytoplasmic assembly, while the C-terminus (~500-1000 amino acid residues) comprises the transmembrane regions (Figure 2). The precise number of transmembrane segments per single monomer unit is controversial; four-, six-, and ten-segment models have been proposed (20). However, a developing consensus supports that six transmembrane segments exist per monomer, in a fashion similar to that of IP3R (88).
RyRs are regulated by numerous natural ligands. Most of these ligands modulate specific sites on the large cytoplasmic portion of the channel. Ligands that activate RyR opening include cytosolic Ca2+, which is the most potent activator, calmodulin (during low Ca2+ concentrations) (110), cyclic ADP ribose (cADPR; primarily an activator of RyR2) (58, 71), ATP (91), and the dihydropyridine receptor (DHPR; for RyR1) (11). Ligand interactions that inhibit the RyR include magnesium (68), FK506 binding protein 12 (FKBP12; for RyR1), and FKBP12.6 (for RyR2) (48). The role of NAADP as a direct allosteric activator of the RyR (versus a two pore channel or a transient receptor potential channel) is still a matter of controversy (13, 28, 56). A list of RyR activators and inhibitors is provided in Table 1.
In addition, protein-protein interactions via three leucine/isoleucine zipper motifs on the RyR2 allow binding of the adaptor proteins spinophilin, PR130, mAKAP, protein phosphatases PP1 and PP2A, and protein kinase A (PKA) to the channel complex (Figure 3) (66).
1.4 Post-translational modifications
In addition to regulation by ligand binding, RyR opening is controlled by post-translational modifications that include covalent phosphorylation, nitrosylation, and redox potential (oxidation/reduction of cysteine sulfhydryl moieties). Three major phosphorylation sites exist on the RyR and physiologically regulate RyR conductance. They include serine (S)2814, S2030, and S2808. S2814 and S2030 are phosphorylated by CaMKII and PKA, respectively. It is not yet resolved which of the kinases-- CaMKII, PKA, or PKG-- phosphorylates S2808 on RyR2.
Increasing evidence supports the redox modulation of RyR channels (44, 84, 112). A small number of hyper-reactive sulfhydryls on RyR are susceptible to oxidation (40), S-nitrosylation (5, 22), and S-glutathionylation (3) by a number of endogenous redox-active agents. Typically, exposure of RyR to these agents increases its sensitivity towards activators or decreases its sensitivity towards inhibitors (3, 75).

Figure 1. Three-dimensional reconstruction of the RyR1 from skeletal muscle by cryo-electron microscopy. Ryanodine receptors were purified from skeletal muscle membrane fractions and cryo-electron microscopy (EM) was performed as previously described (1). (A) “Bottom” view of the surface facing the ER lumen. (B) “Top” view of the cytoplasmic surface. Single arrow represents a cluster of domains that form the cytoplasmic assembly, termed “clamps.” Double arrows represent the largest cytoplasmic assembly domains, which connect the “clamps”, termed “handles.” (C) “Side” view of the transmembrane assembly. Reprinted with permission from Macmillan Publishers Ltd: Nature Structural Biololgy, (93).
1.5 The RyR pore
The RyR pore is the region which provides a pathway for ions to cross the dielectric barrier of the ER membrane and shares much of its architecture with K+ channels. The putative membrane-spanning regions, which make up the RyR pore, have been identified within the last 1000 residues of the C-terminus (20, 104, 127). The ion handling properties of the RyR pore have also been examined in detailed (111). Although the RyR effectively excludes anions, it does not discriminate between cations as well. RyR channels are permeable to a wide range of divalent and monovalent inorganic cations and some organic monovalent cations. The rate of cation translocation through the RyR pore is higher than those of other ion channels (111). Because the RyR behaves as a single ion channel, the most likely explanation is that the RyR pore contains a short, wide, selectivity filter.
1.6 Ca2+-induced Ca2+ release
In skeletal muscle, physical coupling of RyR1 with the DHPR causes RyR opening. However, in cardiac, smooth muscle, and non-excitable cells, RyR Ca2+ release is due to exposure of the RyR to cytosolic Ca2+. This phenomenon is known as Ca2+-induced Ca2+ release (CICR). The RyR is, in fact, a prototypic CICR channel. This triggering Ca2+ signal can arise from several different sources, including extracellular Ca2+ influx through the plasma membrane or intracellular Ca2+ released from neighboring IP3Rs. Regardless of the source, the effectiveness of the Ca2+ signal is dependent on both its speed and amplitude. Work by Fabiato and colleagues demonstrated that SR Ca2+ release is directly proportional to the amplitude and duration of the initial Ca2+ trigger. CICR subsequently undergoes feedback inhibition through binding of Ca2+ to low affinity Ca2+ binding sites on the RyR that are inactivating (8, 23).
1.7 RyR-associated pathologies
Over 300 RyR mutations have been associated with disorders (54), including the RyR myopathies malignant hyperthermia (69, 72), central core disease (63), multiminicore disease (25), atypical periodic paralyses (125), catecholaminergic polymorphic ventricular tachycardia, and arrhythmogenic right ventricular dysplasia type 2 (64, 87, 120). The RyR has also been indirectly linked to acquired pathologies such as heart failure (95) and over-exercise muscle fatigue (4).
The following references provide a more comprehensive review of RyR structure and function (26, 39, 54, 90, 118, 126).

Figure 2. Structural domains and accessory proteins of the RyR2. The primary structure of the RyR and binding domains of protein phosphatases 1 and 2A (PP1, PP2A), protein kinase A (PKA), calmodulin, and FKBP12.6 are shown. PP1 and PP2A and PKA bind to the RyR via their specific adaptor proteins. Six transmembrane segments are shown as previously described (20). CaM, calmodulin; DR, divergent region; FKBP, calstabin-2; LIZ, leucine–isoleucine zipper; SR, sarcoplasmic reticulum. Reprinted with permission from Macmillan Publishers Ltd: Nature Clinical Practice Cardiovascular Medicine, (116).
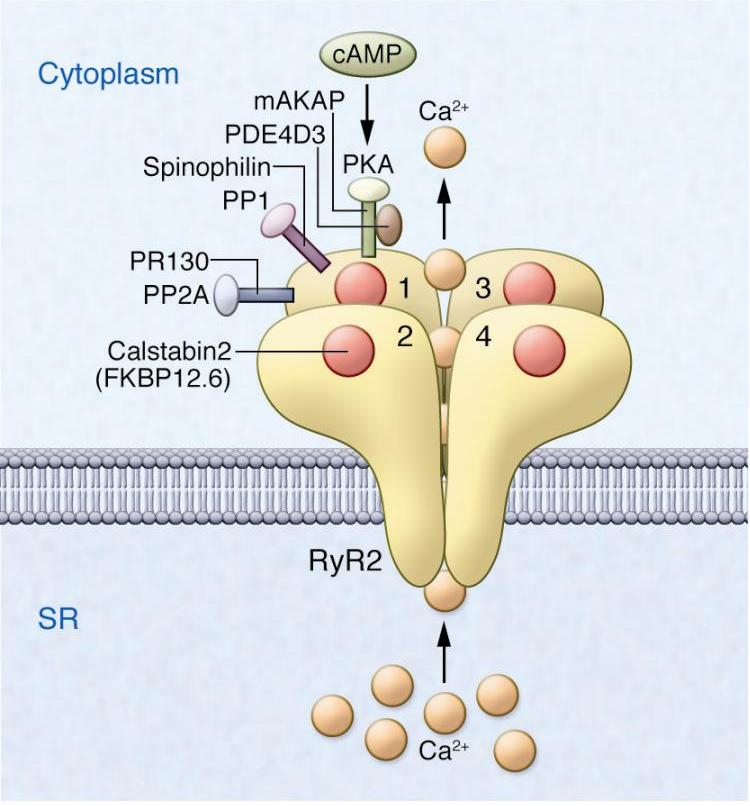
Figure 3. Diagram of the yR2 macromolecular complex with accessory proteins. The RyR2 macromolecular complex includes four identical RyR2 subunits (numerals 1-4). Each RyR2 subunit binds one calstabin2 (also known as FKBP12.6) as well as mAKAP, to which PKA catalytic and regulatory subunits and PDE4D3 are bound; PP2A and its targeting protein PR130; and PP1 and its targeting protein spinophilin (accessory molecules are only shown for one of the four RyR2 subunits, except calstabin2, which is shown for all four RyR2 subunits). The β-adrenergic signaling pathway can activate PKA through the second messenger cAMP. Reprinted with permission from the American Society for Clinical Investigation: Journal of Clinical Investigation, (65).
2. Pancreatic Information
2.1 RyR expression in the pancreas
Several groups have examined RyR isoforms in both acinar cells and islets. Muallem and colleagues failed to see expression of RyRs in rat pancreatic acini by immunofluorescence (IF) (57). Later, however, Nathanson and colleagues demonstrated expression of RyR2 by PCR and western blotting (59). By IF, they saw a non-apical pattern of labeling. A similar distribution of the RyR signal was observed by several labs including those of Yule (using BODIPY-ryanodine) (100) and Pandol (27). The latter found that all three isoforms were expressed. Our lab has further confirmed the presence of RyR in rat (Figure 4) (46, 94), mouse (Figure 5) (78), and in human acinar cells (unpublished).
The presence of RyR in pancreatic islets has been a subject of recent interest. Investigators first considered the possibility that β-cells possess RyR channels after discovering that IP3 caused release of only ~50% of the ER Ca2+ pool (98). Intracellular Ca2+ release is critical for physiologic regulation of insulin secretion by glucose and incretin hormones in pancreatic β-cells (47). Therefore, it was hypothesized that the β-cell ER may be equipped with RyR channels to help regulate this process. Initial studies utilized the RyR activator thimerosal to show intracellular release of Ca2+ from the IP3-insensitive pool (41, 122). The release was potentiated by caffeine, suggesting that RyRs are present. Subsequent studies demonstrated that low dose ryanodine-induced Ca2+ release from islet microsomes (31) and caffeine-induced Ca2+ release from the ER of intact β-cells (80). Work by Johnson et. al. first demonstrated the predominance of RyR2 in mouse islets (6, 47, 50). These studies were recently complemented with the interesting finding that a novel RyR2 splice variant exists in mouse pancreatic islets (107). The protein product is present in both mouse and human islets and remains fully functional. Lastly, Wehrens and colleagues confirmed the original findings that RyR2 is the predominant isoform, with the addition that RyR3 is also expressed at low levels (6).
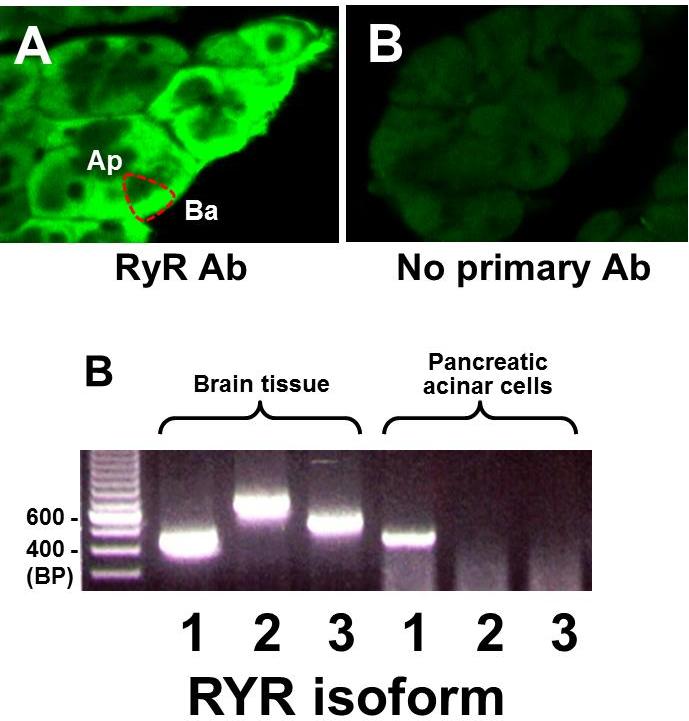
Figure 4. In mouse pancreatic acinar cells the ryanodine receptor (RYR) is localized to the basolateral region, and RYR1 is expressed. (A) Left: RYR is distributed in the basolateral region (Ba) of the acinar cell but excluded from the apical region (Ap). Right: control section without primary antibody but same laser confocal microscopy setting as at left. (B) PCR for RYR isoforms from brain tissue (positive control) and acinar cells. Reprinted with permission from the American Physiological Society: American Journal of Physiology: Gastrointestinal and Liver Physiology, (78).
2.2 RyR Ca2+ release in the acinar cell
RyR Ca2+ release in pancreatic acinar cells was first implicated with the use of caffeine (109). In these early studies, Petersen and colleagues described the globalization of acinar cell Ca2+ signals in the presence of 1 mM caffeine. These results suggested the presence of a RyR releasable Ca2+ pool that is triggered by IP3R-Ca2+ release. Later, Thorn et. al. described a primary role for the RyR in the generation of Ca2+ spikes (105). Even though RyRs appear to be concentrated in the non-apical region of acinar cells (as mentioned earlier), there is some controversy, over whether they can shape apical
Ca2+ signals or whether they primarily influence apical to basal Ca2+ waves. Evidence for both sides will be briefly presented below.
Thorn et. al. demonstrated that intracellular injection of the putative RyR ligand cADPR via a patch pipette induced apical Ca2+ spikes (105). This and other works suggested the presence of RyRs within acidic, non-ER, Ca2+ pools along the apical pole (35, 86). Leite et. al., however, showed that local uncaging of cADPR in the apical region failed to induce a Ca2+ transient, as opposed to uncaging into the basal region (58). RyRs have been implicated as crucial factors in the generation of agonist-induced apical to basal Ca2+ waves. Ryanodine at micromolar concentrations that inhibit CICR markedly reduced the speed of the Ca2+ wave (74, 100). In summary, RyRs are concentrated in the non-apical region, and they amplify Ca2+ signals that are initiated by apical IP3Rs.
2.3 The RyR in pancreas pathology
Whereas physiologic stimuli provoke low amplitude, oscillatory Ca2+ signals in acinar cells, pathologic insults (which cause pancreatic injury and pancreatitis in vivo) induce high amplitude, non-oscillatory Ca2+ signals. These latter aberrant Ca2+ signals are associated with acinar pathology such as intra-acinar protease activation (53, 89), vacuole formation (97), mitochondrial depolarization (89), and acinar cell leakage (45).
Husain et. al. reported that RyRs modulate CCK- and carbachol-induced protease activation (46). Dantrolene, the RyR inhibitor used in this study, appeared to selectively reduce basal Ca2+ signals. Notably, neither dantrolene nor micromolar ryanodine (the latter from a previous study (74)) affected enzyme secretion. The findings implicate the RyR as a potential therapeutic for pancreatitis without the side-effect of perturbing pancreatic function (46, 78).
Ethanol, a major etiology of pancreatitis, given at concentrations that are achievable during intoxication (100 mM) accelerated the apical to basal Ca2+ wave generated by carbachol (79). Ethanol also enhanced intra-acinar protease activation and acinar cell injury. These changes were dependent on the RyR (Figure 6). Another study confirmed that RyR inhibition (using ruthenium red) modulated pathological Ca2+ release and protease activation due to ethanol exposure in permeabilized acinar cells (33). Although the mechanism by which ethanol sensitizes RyRs is still unclear, a potential target is post-translational modifications by PKA phosphorylation (94).
Ethanol is readily converted by the pancreas to its non-oxidative metabolites in the presence of fatty acids to fatty acid ethyl esters (55). Whereas the oxidative metabolites induce mild Ca2+ signals, several of the non-oxidative metabolites including palmitoleic acid ethyl ester (POAEE) and ethyl palmitate both induce strong Ca2+ signals and convert oscillatory signals with physiological stimuli to that of a pathologic peak-plateau pattern (16, 17, 19). The Ca2+ signals with POAEE were partially dependent on the RyR (34). Bile acid exposure of acinar cells may constitute an important mechanism of biliary pancreatitis (82, 83). The bile acid taurolithocholic acid 3-sulfate (TLCS) induces robust peak-plateau Ca2+ signals (32, 108) and acinar cell injury that are dependent upon the RyR (45). Further, in vivo pancreatitis due to infusion of bile acids (TLCS or taurocholic acid; TC) can be prevented and, importantly, treated with the RyR inhibitor dantrolene (45). These data do not negate the potential involvement of IP3Rs that are concentrated in the apical region but also sparsely distributed along the ER in the basal region of the acinar cell. Overall, however, the findings provide potential for the use of RyR modulators as an adjunctive therapy for pancreatitis.
Recent studies have also examined the importance of the RyR in pancreatic islets. It has previously been established that the RyR serves an important function in regulating Ca2+ handling, insulin secretion, and glucose tolerance in pancreatic islets (6, 50).
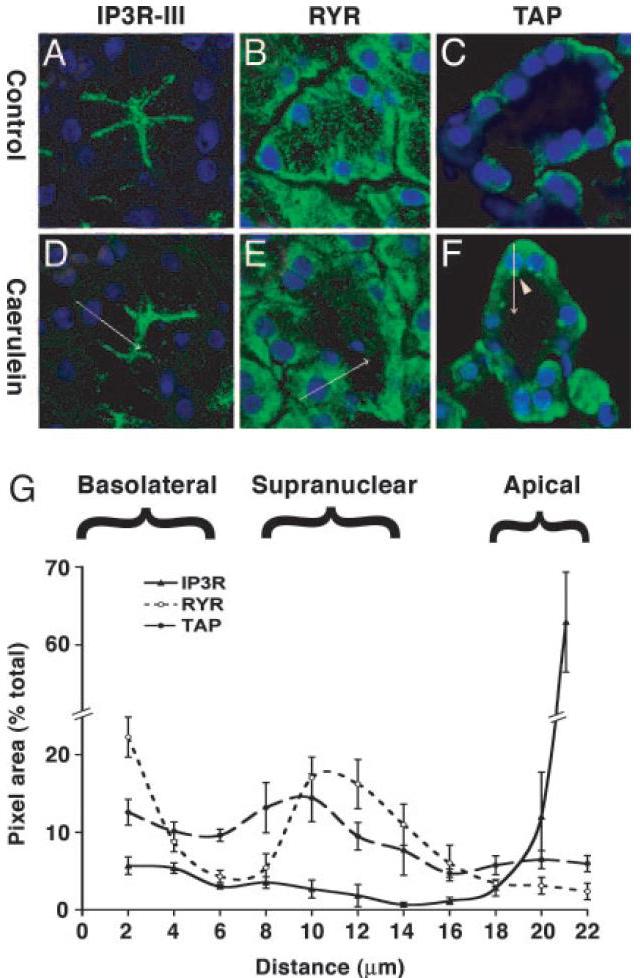
Figure 5. The distribution of Trypsin Activation Peptide (TAP) overlaps with RYR but not with IP3R. Confocal microscopy images of pancreas sections after 30 min of normal saline (A–C) or caerulein hyperstimulation (D–F) in vivo were labeled for IP3R-III (A and D), RYR (B and E), or TAP (C and F); nuclei were stained blue with TOPRO-3. Basal-to-apical line scans (see arrows in D–F) show that IP3R labeling is apical; RYR is distributed in the basolateral region, concentrated in the supranuclear region but excluded from the apical region. TAP appears as discrete supranuclear structures (arrowhead). (G) Overlap between TAP and RYR, but not IP3R, in the supranuclear region is quantified in five line scans from each section, relative to the distance from the basolateral membrane with the nucleus as a reference point. Reprinted with permission from the United States National Academy of Sciences: Proceedings of the National Academy of Sciences, (46).
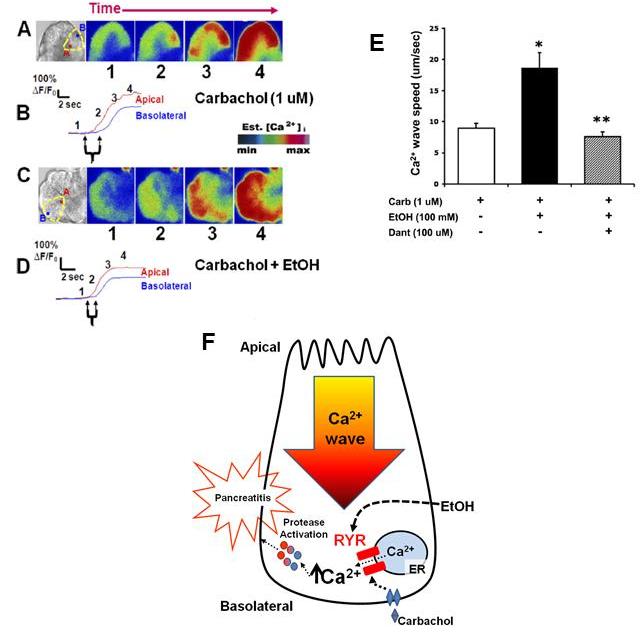
Figure 6. Ethanol accelerates the physiologic carbachol-stimulated Ca2+ wave. (A) Acinar cells were treated with or without ethanol (100 mM) for 30min prior to carbachol (1 uM) stimulation. From left to right, bright field view of an acinus labeled at the apical (A) and basolateral (B) regions of interest from an acinar cell. Cells were loaded with the Ca2+ indicator fluo-4 (5 uM). Upon stimulation with physiologic carbachol (1 uM), subsequent images show the initiation of the Ca2+ signal in the apical region followed by propagation to the basal region. (B) Each paneled image (1– 4), corresponds to a frame along a representative tracing of change in fluorescence over time for each region of interest. Left and right arrows show time of first Ca2+ rise in the apical and basal regions, respectively. Est. [Ca2+]i, estimated [Ca2+]i; min, minimum; max, maximum. (C-D) Cells were pretreated with ethanol (100 mM) for 30 min. (E) Quantitation of difference in Ca2+ wave speed with carbachol + ethanol in the presence or absence of dantrolene (Dant, 100 uM). (n =13 cells in each). *, p<0.005. (F) Proposed mechanism by which ethanol evokes pathological effects on the pancreatic acinar cell. Reprinted with permission from the American Society for Biochemistry and Molecular Biology: Journal of Biological Chemistry, (79).
3. Tools for Study
3.1 Antibodies
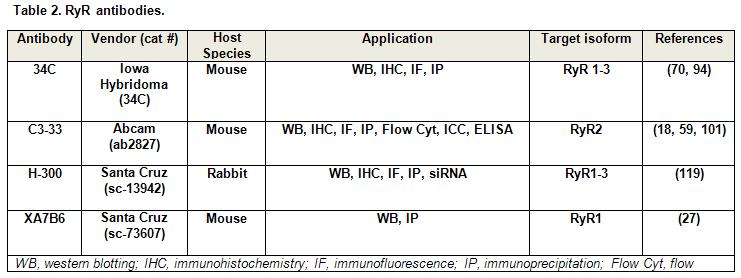
A list of RyR antibodies and their applications are provided in Table 2.
3.2 Expression vectors
RyR1 and RyR2 have been overexpressed in HEK293 cells by Chen and colleagues (49). These constructs have proven useful for examining the role of various mutations on RyR Ca2+ release.
3.3 Animal models
The RyR1 (102) and RyR2 (103) knockout mice do not survive beyond neonatal life, while RyR3 deficient mice live to adulthood (7). Currently, there are no floxed conditional knockouts. There is, however, a knockout of CD38, the enzyme that synthesizes the putative RyR activator cADPR (77, 117). There are also knockins of the phosphomimetic and phosphoresistant mutations at S2808 and S2814 on RyR2 (95, 96). A certain strain of pig has a higher incidence of RyR mutations that predispose them to malignant hyperthermia (99). A knockin mouse model of malignant hyperthermia has also been established (24). .
3.4 Ligand binding
Ryanodine binding studies are useful to examine (1) the presence of RyRs and (2) the density of RyRs in cells or tissues of interest and (3) to indirectly determine RyR open state. A detailed methods protocol for performing tritiated ryanodine binding is provided by Meissner and colleagues (21)
3.4 Activators and inhibitors
A list of RyR activators and inhibitors are in Table 1.
3.5 Pull-down
Pull-down of RyR is achieved with high affinity using a FKBP12 or FKBP12.6 GST fusion protein (51, 61).

References
- Akey CW, Radermacher M. Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J Cell Biol 122: 1-19, 1993. PMID: 8314837
- Allen PD, Lopez JR, Sanchez V, Ryan JF, Sreter FA. EU 4093 decreases intracellular [Ca2+] in skeletal muscle fibers from control and malignant hyperthermia-susceptible swine. Anesthesiology 76: 132-138, 1992 PMID: 1729917
- Aracena P, Sanchez G, Donoso P, Hamilton SL, Hidalgo C. S-glutathionylation decreases Mg2+ inhibition and S-nitrosylation enhances Ca2+ activation of RyR1 channels. J Biol Chem 278: 42927-42935, 2003. PMID: 12920114
- Bellinger AM, Mongillo M, Marks AR. Stressed out: the skeletal muscle ryanodine receptor as a target of stress. Clin Invest 118: 445-453, 2008. PMID: 18246195
- Bellinger AM, Reiken S, Carlson C, Mongillo M, Liu X, Rothman L, Matecki S, Lacampagne A, Marks AR. Hypernitrosylated ryanodine receptor calcium release channels are leaky in dystrophic muscle. Nat Med 15: 325-330, 2009. PMID: 19198614
- Bennett DL, Cheek TR, Berridge MJ, De Smedt H, Parys JB, Missiaen L, Bootman MD. Expression and function of ryanodine receptors in nonexcitable cells. J Biol Chem 271: 6356-6362, 1996. PMID: 8626432
- Bertocchini F, Ovitt CE, Conti A, Barone V, Scholer HR, Bottinelli R, Reggiani C, and Sorrentino V. Requirement for the ryanodine receptor type 3 for efficient contraction in neonatal skeletal muscles. Embo J 16: 6956-6963, 1997. PMID: 9384575
- Bezprozvanny I, Watras J, Ehrlich BE. Bell-shaped calcium-response curves of Ins(1,4,5)P3- and calcium-gated channels from endoplasmic reticulum of cerebellum. Nature 351: 751-754, 1991. PMID: 1648178
- Bidasee KR, Besch HR, Jr. Structure-function relationships among ryanodine derivatives. Pyridyl ryanodine definitively separates activation potency from high affinity. J Biol Chem 273: 12176-12186, 1998. PMID: 9575165
- Bina S, Cowan G, Karaian J, Muldoon S, Mongan P, Bunger R. Effects of caffeine, halothane, and 4-chloro-m-cresol on skeletal muscle lactate and pyruvate in malignant hyperthermia-susceptible and normal swine as assessed by microdialysis. Anesthesiology 104: 90-100, 2006. PMID: 16394695
- Block BA, Imagawa T, Campbell KP, Franzini-Armstrong C. Structural evidence for direct interaction between the molecular components of the transverse tubule/sarcoplasmic reticulum junction in skeletal muscle. J Cell Biol 107: 2587-2600, 1988. PMID: 2849609
- Bradley KN, Currie S, MacMillan D, Muir TC, McCarron JG. Cyclic ADP-ribose increases Ca2+ removal in smooth muscle. J Cell Sci 116: 4291-4306, 2003. PMID: 12966165
- Cang C, Zhou Y, Navarro B, Seo YJ, Aranda K, Shi L, Battaglia-Hsu S, Nissim I, Clapham DE, Ren D. mTOR regulates lysosomal ATP-sensitive two-pore Na(+) channels to adapt to metabolic state. Cell 152: 778-790, 2013. PMID: 23394946
- Choi KJ, Kim KS, Kim SH, Kim DK, Park HS. Caffeine and 2-Aminoethoxydiphenyl Borate (2-APB) Have Different Ability to Inhibit Intracellular Calcium Mobilization in Pancreatic Acinar Cell. Korean J Physiol Pharmacol 14: 105-111, 2010. PMID: 20473382
- Chu A, Diaz-Munoz M, Hawkes MJ, Brush K, Hamilton SL. Ryanodine as a probe for the functional state of the skeletal muscle sarcoplasmic reticulum calcium release channel. Mol Pharmacol 37: 735-741, 1990. PMID: 1692609
- Criddle DN, Murphy J, Fistetto G, Barrow S, Tepikin AV, Neoptolemos JP, Sutton R, Petersen OH. Fatty acid ethyl esters cause pancreatic calcium toxicity via inositol trisphosphate receptors and loss of ATP synthesis. Gastroenterology 130: 781-793, 2006. PMID: 16530519
- Criddle DN, Raraty MG, Neoptolemos JP, Tepikin AV, Petersen OH, Sutton R. Ethanol toxicity in pancreatic acinar cells: mediation by nonoxidative fatty acid metabolites. Proc Natl Acad Sci U S A 101: 10738-10743, 2004. PMID: 15247419
- Dixit SS, Wang T, Manzano EJ, Yoo S, Lee J, Chiang DY, Ryan N, Respress JL, Yechoor VK, Wehrens XH. Effects of CaMKII-mediated phosphorylation of ryanodine receptor type 2 on islet calcium handling, insulin secretion, and glucose tolerance. PLoS One 8: e58655, 2013. PMID: 23516528
- Dolai S, Liang T, Lam PP, Fernandez NA, Chidambaram S, and Gaisano HY. Effects of ethanol metabolites on exocytosis of pancreatic acinar cells in rats. Gastroenterology 143: 832-843, 2012. PMID: 22710192
- Du GG, Sandhu B, Khanna VK, Guo XH, MacLennan DH. Topology of the Ca2+ release channel of skeletal muscle sarcoplasmic reticulum (RyR1). Proc Natl Acad Sci U S A 99: 16725-16730, 2002. PMID: 12486242
- Eu JP, Meissner G. Detection of calcium release via ryanodine receptors. Methods Mol Biol 798: 373-382, 2012. PMID: 22130848
- Eu JP, Sun J, Xu L, Stamler JS, Meissner G. The skeletal muscle calcium release channel: coupled O2 sensor and NO signaling functions. Cell 102: 499-509, 2000. PMID: 10966111
- Fabiato A. Effects of ryanodine in skinned cardiac cells. Fed Proc 44: 2970-2976, 1985. PMID: 2415405
- Fairhurst AAS, Hamamoto VV, Macri JJ. Modification of ryanodine toxicity by dantrolene and halothane in a model of malignant hyperthermia. Anesthesiology 53: 199-204, 1980. PMID: 7425333
- Ferreiro A, Monnier N, Romero NB, Leroy JP, Bonnemann C, Haenggeli CA, Straub V, Voss WD, Nivoche Y, Jungbluth H, Lemainque A, Voit T, Lunardi J, Fardeau M, Guicheney P. A recessive form of central core disease, transiently presenting as multi-minicore disease, is associated with a homozygous mutation in the ryanodine receptor type 1 gene. Ann Neurol 51: 750-759, 2002. PMID: 12112081
- Fill M, and Copello JA. Ryanodine receptor calcium release channels. Physiol Rev 82: 893-922, 2002. PMID: 12270947
- Fitzsimmons TJ, Gukovsky I, McRoberts JA, Rodriguez E, Lai FA, Pandol SJ. Multiple isoforms of the ryanodine receptor are expressed in rat pancreatic acinar cells. Biochem J 351: 265-271, 2000. PMID: 10998370
- Fliegert R, Gasser A, Guse AH. Regulation of calcium signalling by adenine-based second messengers. Biochem Soc Trans 35: 109-114, 2007. PMID: 17233614
- Furuichi T, Furutama D, Hakamata Y, Nakai J, Takeshima H, Mikoshiba K. Multiple types of ryanodine receptor/Ca2+ release channels are differentially expressed in rabbit brain. J Neurosci 14: 4794-4805, 1994. PMID: 8046450
- Futatsugi A, Kuwajima G, Mikoshiba K. Tissue-specific and developmentally regulated alternative splicing in mouse skeletal muscle ryanodine receptor mRNA. Biochem J 305 ( Pt 2): 373-378, 1995. PMID: 7832748
- Gao L, Balshaw D, Xu L, Tripathy A, Xin C, Meissner G. Evidence for a role of the lumenal M3-M4 loop in skeletal muscle Ca(2+) release channel (ryanodine receptor) activity and conductance. Biophys J 79: 828-840, 2000. PMID: 10920015
- Gerasimenko JV, Flowerdew SE, Voronina SG, Sukhomlin TK, Tepikin AV, Petersen OH, Gerasimenko OV. Bile acids induce Ca2+ release from both the endoplasmic reticulum and acidic intracellular calcium stores through activation of inositol trisphosphate receptors and ryanodine receptors. J Biol Chem 281: 40154-40163, 2006. PMID: 17074764
- Gerasimenko JV, Lur G, Ferdek P, Sherwood MW, Ebisui E, Tepikin AV, Mikoshiba K, Petersen OH, Gerasimenko OV. Calmodulin protects against alcohol-induced pancreatic trypsinogen activation elicited via Ca2+ release through IP3 receptors. Proc Natl Acad Sci U S A 108: 5873-5878, 2011. PMID: 21436055
- Gerasimenko JV, Lur G, Sherwood MW, Ebisui E, Tepikin AV, Mikoshiba K, Gerasimenko OV, Petersen OH. Pancreatic protease activation by alcohol metabolite depends on Ca2+ release via acid store IP3 receptors. Proc Natl Acad Sci U S A 106: 10758-10763, 2009. PMID: 19528657
- Gerasimenko JV, Sherwood M, Tepikin AV, Petersen OH, Gerasimenko OV. NAADP, cADPR and IP3 all release Ca2+ from the endoplasmic reticulum and an acidic store in the secretory granule area. J Cell Sci 119: 226-238, 2006. PMID: 16410548
- Giannini G, Conti A, Mammarella S, Scrobogna M, Sorrentino V. The ryanodine receptor/calcium channel genes are widely and differentially expressed in murine brain and peripheral tissues. J Cell Biol 128: 893-904, 1995. PMID: 7876312
- Grunwald R, Meissner G. Lumenal sites and C terminus accessibility of the skeletal muscle calcium release channel (ryanodine receptor). J Biol Chem 270: 11338-11347, 1995. PMID: 7744771
- Gu X, Yang Z, Zhang L, Kunerth S, Fliegert R, Weber K, Guse AH. Synthesis and biological evaluation of novel membrane-permeant cyclic ADP-ribose mimics: N1-[(5''-O-phosphorylethoxy)methyl]-5'-O-phosphorylinosine 5',5''-cyclicpyrophosphate (cIDPRE) and 8-substituted derivatives. J Med Chem 47: 5674-5682, 2004. PMID: 15509166
- Guida L, Bruzzone S, Sturla L, Franco L, Zocchi E, De Flora A. Equilibrative and concentrative nucleoside transporters mediate influx of extracellular cyclic ADP-ribose into 3T3 murine fibroblasts. J Biol Chem 277: 47097-47105, 2002. PMID: 12368285
- Hamilton SL, Reid MB. RyR1 modulation by oxidation and calmodulin. Antioxid Redox Signal 2: 41-45, 2000. PMID: 11232598
- Harteneck C, Plant TD, Schultz G. From worm to man: three subfamilies of TRP channels. Trends Neurosci 23: 159-166, 2000. PMID: 10717675
- Hermann-Frank A, Richter M, Lehmann-Horn F. 4-Chloro-m-cresol: A specific tool to distinguish between malignant hyperthermia-susceptible and normal muscle. Biochem Pharmacol 52: 149-155, 1996. PMID: 8678899
- Herrmann-Frank A, Richter M, Sarközi S, Mohr U, Lehmann-Horn F. 4-chloro-m-cresol, a potent and specific activator of the skeletal muscle ryanodine receptor. Biochim et Biophys Acta (BBA) - General Subjects 1289: 31-40, 1996. PMID: 8605229
- Hidalgo C, Aracena P, Sanchez G, Donoso P. Redox regulation of calcium release in skeletal and cardiac muscle. Biol Res 35: 183-193, 2002. PMID: 12415735
- Husain SZ, Orabi AI, Muili KA, Luo Y, Sarwar S, Mahmood SM, Wang D, Choo-Wing R, Singh VP, Parness J, Ananthanaravanan M, Bhandari V, Perides G. Ryanodine receptors contribute to bile acid-induced pathological calcium signaling and pancreatitis in mice. Am J Physiol: Gastrointest Liver Physiol 302: G1423-1433, 2012. PMID: 22517774
- Husain SZ, Prasad P, Grant WM, Kolodecik TR, Nathanson MH, Gorelick FS. The ryanodine receptor mediates early zymogen activation in pancreatitis. Proc Natl Acad Sci U S A 102: 14386-14391, 2005. PMID: 16186498
- Islam MS. The Ryanodine Receptor Calcium Channel of {beta}-Cells: Molecular Regulation and Physiological Significance. Diabetes 51: 1299-1309, 2002. PMID: 11978625
- Jayaraman T, Brillantes A, Timerman A, Fleischer S, Erdjument-Bromage H, Tempst P, Marks A. FK506 binding protein associated with the calcium release channel (ryanodine receptor). J Biol Chem 267: 9474-9477, 1992. PMID: 1374404
- Jiang D, Xiao B, Li X, Chen SRW. Smooth Muscle Tissues Express a Major Dominant Negative Splice Variant of the Type 3 Ca2+ Release Channel (Ryanodine Receptor). J Biol Chem 278: 4763-4769, 2003. PMID: 12471029
- Johnson JD, Kuang S, Misler S, Polonsky KS. Ryanodine receptors in human pancreatic β cells: localization and effects on insulin secretion. FASEB J 03-1280fje, 2004. PMID: 15033925
- Jones PP, Meng X, Xiao B, Cai S, Bolstad J, Wagenknecht T, Liu Z, Chen SRW. Localization of PKA phosphorylation site, Ser2030, in the three-dimensional structure of cardiac ryanodine receptor. Biochem J 410: 261-270, 2008. PMID: 17967164
- Kobayashi S, Bannister ML, Gangopadhyay JP, Hamada T, Parness J, Ikemoto N. Dantrolene stabilizes domain interactions within the Ryanodine receptor. J Biol Chem 2004. PMID: 15611117
- Kruger B, Albrecht E, Lerch MM. The role of intracellular calcium signaling in premature protease activation and the onset of pancreatitis. Am J Pathol 157: 43-50, 2000. PMID: 10880374
- Lanner JT, Georgiou DK, Joshi AD, Hamilton SL. Ryanodine receptors: structure, expression, molecular details, and function in calcium release. Cold Spring Harb on Perspect Biol 2: a003996, 2010. PMID: 20961976
- Laposata EA, Lange LG. Presence of nonoxidative ethanol metabolism in human organs commonly damaged by ethanol abuse. Science 231: 497-499, 1986. PMID: 3941913
- Lee HC. Physiological functions of cyclic ADP-ribose and NAADP as calcium messengers. Annu Rev Pharmacol Toxicol 41: 317-345, 2001. PMID: 11264460
- Lee MG, Xu X, Zeng W, Diaz J, Wojcikiewicz RJ, Kuo TH, Wuytack F, Racymaekers L, Muallem S. Polarized expression of Ca2+ channels in pancreatic and salivary gland cells. Correlation with initiation and propagation of [Ca2+]i waves. J Biol Chem 272: 15765-15770, 1997. PMID: 9188472
- Leite MF, Burgstahler AD, Nathanson MH. Ca2+ waves require sequential activation of inositol trisphosphate receptors and ryanodine receptors in pancreatic acini. Gastroenterology 122: 415-427, 2002. PMID: 11832456
- Leite MF, Dranoff JA, Gao L, Nathanson MH. Expression and subcellular localization of the ryanodine receptor in rat pancreatic acinar cells. Biochem J 337 ( Pt 2): 305-309, 1999. PMID: 9882629
- Leslie GC, Part NJ. The effect of EU4093 (azumolene sodium) on the contraction of intrafusal muscle in the soleus muscle of the anaesthetized rat. Br J Pharmacol 97: 1151-1156, 1989. PMID: 2790379
- Liu Z, Zhang J, Wang R, Wayne Chen SR, Wagenknecht T. Location of divergent region 2 on the three-dimensional structure of cardiac muscle ryanodine receptor/calcium release channel. J Mol Biol 338: 533-545, 2004. PMID: 15081811
- Lobo PA, and Van Petegem F. Crystal structures of the N-terminal domains of cardiac and skeletal muscle ryanodine receptors: insights into disease mutations. Structure 17: 1505-1514, 2009. PMID: 19913485
- Lynch PJ, Tong J, Lehane M, Mallet A, Giblin L, Heffron JJ, Vaughan P, Zafra G, MacLennan DH, McCarthy TV. A mutation in the transmembrane/luminal domain of the ryanodine receptor is associated with abnormal Ca2+ release channel function and severe central core disease. Proc Natl Acad Sci U S A 96: 4164-4169, 1999. PMID: 10097181
- Magee KR, Shy GM. A new congenital non-progressive myopathy. Brain 79: 610-621, 1956. PMID: 13396066
- Marks AR. Calcium cycling proteins and heart failure: mechanisms and therapeutics. J Clin Invest 123: 46-52, 2013. PMID: 23281409
- Marx SO, Reiken S, Hisamatsu Y, Jayaraman T, Burkhoff D, Rosemblit N, Marks AR. PKA Phosphorylation Dissociates FKBP12.6 from the Calcium Release Channel (Ryanodine Receptor): Defective Regulation in Failing Hearts. Cell 101: 365-376, 2000. PMID:10830164
- McPherson PS, Campbell KP. The ryanodine receptor/Ca2+ release channel. J Biol Chem 268: 13765-13768, 1993. PMID: 8390976
- Meissner G, Darling E, Eveleth J. Kinetics of rapid Ca2+ release by sarcoplasmic reticulum. Effects of Ca2+, Mg2+, and adenine nucleotides. Biochemistry 25: 236-244, 1986. PMID: 3754147
- Melzer W, Dietze B. Malignant hyperthermia and excitation-contraction coupling. Acta Physiol Scand 171: 367-378, 2001. PMID: 11412150
- Meng X, Xiao B, Cai S, Huang X, Li F, Bolstad J, Trujillo R, Airey J, Chen SRW, Wagenknecht T, Liu Z. Three-Dimensional Localization of Serine 2808, a Phosphorylation Site in Cardiac Ryanodine Receptor. J Biol Chem 282: 25929-25939, 2007. PMID: 17606610
- Meszaros LG, Bak J, and Chu A. Cyclic ADP-ribose as an endogenous regulator of the non-skeletal type ryanodine receptor Ca2+ channel. Nature 364: 76-79, 1993. PMID: 8391127
- Migita T, Mukaida K, Hamada H, Yasuda T, Haraki T, Nishino I, Murakami N, and Kawamoto M. Functional analysis of ryanodine receptor type 1 p.R2508C mutation in exon 47. J Anesth 23: 341-346, 2009. PMID: 19685112
- Nakai J, Imagawa T, Hakamat Y, Shigekawa M, Takeshima H, Numa S. Primary structure and functional expression from cDNA of the cardiac ryanodine receptor/calcium release channel. FEBS Lett 271: 169-177, 1990. PMID: 2226801
- Nathanson MH, Padfield PJ, O'Sullivan AJ, Burgstahler AD, Jamieson JD. Mechanism of Ca2+ wave propagation in pancreatic acinar cells. J Biol Chem 267: 18118-18121, 1992. PMID: 1517244
- Oba T, Murayama T, Ogawa Y. Redox states of type 1 ryanodine receptor alter Ca(2+) release channel response to modulators. Am J Physiol: Cell Physiol 282: C684-C692, 2002. PMID: 11880257
- Ogunbayo OA, Zhu Y, Rossi D, Sorrentino V, Ma J, Zhu MX, Evans AM. Cyclic adenosine diphosphate ribose activates ryanodine receptors, whereas NAADP activates two-pore domain channels. J Biol Chem 286: 9136-9140, 2011. PMID: 21216967
- Orabi AI, Muili KA, Javed TA, Jin S, Jayaraman T, Lund FE, Husain SZ. Cluster of differentiation 38 (CD38) mediates bile-acid induced acinar cell injury and pancreatitis through cyclic ADP ribose and intracellular calcium release. J Biol Chem 288: 27128-27137, 2013. PMID: 23940051
- Orabi AI, Shah AU, Ahmad MU, Choo-Wing R, Parness J, Jain D, Bhandari V, Husain SZ. Dantrolene mitigates caerulein-induced pancreatitis in vivo in mice. Am J Physiol: Gastrointest Liver Physiol 299: G196-204, 2009. PMID: 20448143
- Orabi AI, Shah AU, Muili K, Luo Y, Mahmood SM, Ahmad A, Reed A, Husain SZ. Ethanol enhances carbachol-induced protease activation and accelerates Ca2+ waves in isolated rat pancreatic acini. J Biol Chem 286: 14090-14097, 2011. PMID: 21372126
- Oyamada H, Murayama T, Takagi T, Iino M, Iwabe N, Miyata T, Ogawa Y, Endo M. Primary structure and distribution of ryanodine-binding protein isoforms of the bullfrog skeletal muscle. J Biol Chem 269: 17206-17214, 1994. PMID: 8006029
- Paul-Pletzer K, Yamamoto T, Ikemoto N, Jimenez LS, Morimoto H, Williams PG, Ma J, Parness J. Probing a putative dantrolene binding site on the cardiac ryanodine receptor. Biochem J 2005. PMID: 15656791
- Perides G, Laukkarinen JM, Vassileva G, Steer ML. Biliary acute pancreatitis in mice is mediated by the G-protein-coupled cell surface bile acid receptor Gpbar1. Gastroenterology 138: 715-725, 2010. PMID: 19900448
- Perides G, van Acker GJ, Laukkarinen JM, Steer ML. Experimental acute biliary pancreatitis induced by retrograde infusion of bile acids into the mouse pancreatic duct. Nat Protoc 5: 335-341, 2010. PMID: 20134432
- Pessah IN, Kim KH, Feng W. Redox sensing properties of the ryanodine receptor complex. Front Biosci 7: a72-79, 2002. PMID: 11991848
- Pessah IN, Zimanyi I. Characterization of multiple [3H]ryanodine binding sites on the Ca2+ release channel of sarcoplasmic reticulum from skeletal and cardiac muscle: evidence for a sequential mechanism in ryanodine action. Mol Pharmacol 39: 679-689, 1991. PMID: 1851961
- Petersen OH, Tepikin AV. Polarized calcium signaling in exocrine gland cells. Annu Rev Physiol 70: 273-299, 2008. PMID: 17850212
- Phillips MS, Khanna VK, De Leon S, Frodis W, Britt BA, MacLennan DH. The substitution of Arg for Gly2433 in the human skeletal muscle ryanodine receptor is associated with malignant hyperthermia. Hum Mol Genet 3: 2181-2186, 1994. PMID: 7881417
- Ramachandran S, Chakraborty A, Xu L, Mei Y, Samso M, Dokholyan NV, Meissner G. Structural determinants of skeletal muscle ryanodine receptor gating. J Biol Chem 288: 6154-6165, 2013. PMID: 23319589
- Raraty M, Ward J, Erdemli G, Vaillant C, Neoptolemos JP, Sutton R, Petersen OH. Calcium-dependent enzyme activation and vacuole formation in the apical granular region of pancreatic acinar cells. Proc Natl Acad Sci U S A 97: 13126-13131, 2000. PMID: 11087863
- Rossi D, Sorrentino V. Molecular genetics of ryanodine receptors Ca2+-release channels. Cell Calcium 32: 307-319, 2002. PMID: 12543091
- Rousseau E, Ladine J, Liu QY, Meissner G. Activation of the Ca2+ release channel of skeletal muscle sarcoplasmic reticulum by caffeine and related compounds. Arch Biochem Biophys 267: 75-86, 1988. PMID: 2848455
- Seo MD, Velamakanni S, Ishiyama N, Stathopulos PB, Rossi AM, Khan SA, Dale P, Li C, Ames JB, Ikura M, Taylor CW. Structural and functional conservation of key domains in InsP3 and ryanodine receptors. Nature 483: 108-112, 2012. PMID: 22286060
- Serysheva, II, Orlova EV, Chiu W, Sherman MB, Hamilton SL, van Heel M. Electron cryomicroscopy and angular reconstitution used to visualize the skeletal muscle calcium release channel. Nat Struct Biol 2: 18-24, 1995. PMID: 7719847
- Shah AU, Grant WM, Latif SU, Mannan ZM, Park AJ, Husain SZ. Cyclic-AMP Accelerates Calcium Waves in Pancreatic Acinar Cells. Am J Physiol: Gastrointest Liver Physiol 294: G1328-1334, 2008. PMID: 18388188
- Shan J, Betzenhauser MJ, Kushnir A, Reiken S, Meli AC, Wronska A, Dura M, Chen BX, Marks AR. Role of chronic ryanodine receptor phosphorylation in heart failure and beta-adrenergic receptor blockade in mice. J Clin Invest 120: 4375-4387, 2010. PMID: 21099115
- Shan J, Kushnir A, Betzenhauser MJ, Reiken S, Li J, Lehnart SE, Lindegger N, Mongillo M, Mohler PJ, Marks AR. Phosphorylation of the ryanodine receptor mediates the cardiac fight or flight response in mice. J Clin Invest 120: 4388-4398, 2010. PMID: 21099118
- Sherwood MW, Prior IA, Voronina SG, Barrow SL, Woodsmith JD, Gerasimenko OV, Petersen OH, Tepikin AV. Activation of trypsinogen in large endocytic vacuoles of pancreatic acinar cells. Proc Natl Acad Sci USA 104: 5674-5679, 2007. PMID: 17363470
- Sorrentino V, Barone V, Rossi D. Intracellular Ca(2+) release channels in evolution. Curr Opin Genet Dev 10: 662-667, 2000. PMID: 11088018
- Stadhouders AM, Viering WA, Verburg MP, Ruitenbeek W, Sengers RC. In vivo induced malignant hyperthermia in pigs. III. Localization of calcium in skeletal muscle mitochondria by means of electronmicroscopy and microprobe analysis. Acta Anaesthesiol Scand 28: 14-26, 1984. PMID: 6711259
- Straub SV, Giovannucci DR, Yule DI. Calcium wave propagation in pancreatic acinar cells: functional interaction of inositol 1,4,5-trisphosphate receptors, ryanodine receptors, and mitochondria. J Gen Physiol 116: 547-560, 2000. PMID: 11004204
- Takasawa S, Kuroki M, Nata K, Noguchi N, Ikeda T, Yamauchi A, Ota H, Itaya-Hironaka A, Sakuramoto-Tsuchida S, Takahashi I, Yoshikawa T, Shimosegawa T, Okamoto H. A novel ryanodine receptor expressed in pancreatic islets by alternative splicing from type 2 ryanodine receptor gene. Biochem Biophys Res Commun 397: 140-145, 2010. PMID: 20471962
- Takeshima H, Iino M, Takekura H, Nishi M, Kuno J, Minowa O, Takano H, Noda T. Excitation-contraction uncoupling and muscular degeneration in mice lacking functional skeletal muscle ryanodine-receptor gene. Nature 369: 556-559, 1994. PMID: 7515481
- Takeshima H, Komazaki S, Hirose K, Nishi M, Noda T, Lino M. Embryonic lethality and abnormal cardiac myocytes in mice lacking ryanodine receptor type 2. EMBO J 17: 3309-3316, 1998. PMID: 9628868
- Takeshima H, Nishimura S, Matsumoto T, Ishida H, Kangawa K, Minamino N, Matsuo H, Ueda M, Hanaoka M, Hirose T, and et al. Primary structure and expression from complementary DNA of skeletal muscle ryanodine receptor. Nature 339: 439-445, 1989. PMID: 2725677
- Thorn P, Gerasimenko O, Petersen OH. Cyclic ADP-ribose regulation of ryanodine receptors involved in agonist evoked cytosolic Ca2+ oscillations in pancreatic acinar cells. EMBO J 13: 2038-2043, 1994. PMID: 7514529
- Tunwell RE, Wickenden C, Bertrand BM, Shevchenko VI, Walsh MB, Allen PD, Lai FA. The human cardiac muscle ryanodine receptor-calcium release channel: identification, primary structure and topological analysis. Biochem J 318 ( Pt 2): 477-487, 1996. PMID: 8809036
- Vazquez-Martinez O, Canedo-Merino R, Diaz-Munoz M, Riesgo-Escovar JR. Biochemical characterization, distribution and phylogenetic analysis of Drosophila melanogaster ryanodine and IP3 receptors, and thapsigargin-sensitive Ca2+ ATPase. J Cell Sci 116: 2483-2494, 2003. PMID: 12766186
- Voronina S, Longbottom R, Sutton R, Petersen OH, Tepikin A. Bile acids induce calcium signals in mouse pancreatic acinar cells: implications for bile-induced pancreatic pathology. J Physiol 540: 49-55, 2002. PMID: 11927668
- Wakui M, Osipchuk YV, Petersen OH. Receptor-activated cytoplasmic Ca2+ spiking mediated by inositol trisphosphate is due to Ca2(+)-induced Ca2+ release. Cell 63: 1025-1032, 1990. PMID: 1701691
- Wehrens XH, Lehnart SE, Reiken SR, Marks AR. Ca2+/calmodulin-dependent protein kinase II phosphorylation regulates the cardiac ryanodine receptor. Circ Res 94: e61-70, 2004. PMID: 15016728
- Williams AJ, West DJ, Sitsapesan R. Light at the end of the Ca(2+)-release channel tunnel: structures and mechanisms involved in ion translocation in ryanodine receptor channels. Q Rev Biophys 34: 61-104, 2001. PMID: 11388090
- Xia R, Stangler T, Abramson JJ. Skeletal muscle ryanodine receptor is a redox sensor with a well defined redox potential that is sensitive to channel modulators. J Biol Chem 275: 36556-36561, 2000. PMID: 10952995
- Xu L, Tripathy A, Pasek DA, Meissner G. Potential for pharmacology of ryanodine receptor/calcium release channels. Ann N Y Acad Sci 853: 130-148, 1998. PMID: 10603942
- Xu X, Bhat MB, Nishi M, Takeshima H, Ma J. Molecular cloning of cDNA encoding a drosophila ryanodine receptor and functional studies of the carboxyl-terminal calcium release channel. Biophys J 78: 1270-1281, 2000. PMID: 10692315
- Yamasaki-Mann M, Demuro A, Parker I. Modulation of endoplasmic reticulum Ca2+ store filling by cyclic ADP-ribose promotes inositol trisphosphate (IP3)-evoked Ca2+ signals. J Biol Chem 285: 25053-25061, 2010. PMID: 20538594
- Yano MM, Yamamoto TT, Ikeda YY, Matsuzaki MM. Mechanisms of Disease: ryanodine receptor defects in heart failure and fatal arrhythmia. Nature Clinical Practice Cardiovascular Medicine 3: 43-52, 2006. PMID: 16391617
- Yue J, Wei W, Lam CM, Zhao YJ, Dong M, Zhang LR, Zhang LH, Lee HC. CD38/cADPR/Ca2+ pathway promotes cell proliferation and delays nerve growth factor-induced differentiation in PC12 cells. J Biol Chem 284: 29335-29342, 2009. PMID: 19696022
- Zalk R, Lehnart SE, Marks AR. Modulation of the Ryanodine Receptor and Intracellular Calcium. Ann Rev Biochemistry 76: 2007. PMID:17506640
- Zhang Q, Kohler M, Yang SN, Zhang F, Larsson O, Berggren PO. Growth hormone promotes Ca(2+)-induced Ca2+ release in insulin-secreting cells by ryanodine receptor tyrosine phosphorylation. Mol Endocrinol 18: 1658-1669, 2004. PMID: 15056730
- Zhang Y, Chen HS, Khanna VK, De Leon S, Phillips MS, Schappert K, Britt BA, Browell AK, MacLennan DH. A mutation in the human ryanodine receptor gene associated with central core disease. Nat Genet 5: 46-50, 1993. PMID: 8220422
- Zhang Y, Rodney GG, Schneider MF. Effects of azumolene on Ca2+ sparks in skeletal muscle fibers. J Pharmacol Exp Ther 2005. PMID: 15831441
- Zhao M, Li P, Li X, Zhang L, Winkfein RJ, Chen SR. Molecular identification of the ryanodine receptor pore-forming segment. J Biol Chem 274: 25971-25974, 1999. PMID: 10473538
- Zhao X, Weisleder N, Han X, Pan Z, Parness J, Brotto M, Ma J. Azumolene inhibits a component of store-operated calcium entry coupled to the skeletal muscle ryanodine receptor. J Biol Chem M602306200, 2006. PMID: 16945924
- Zhao Y, Graeff R, Lee HC. Roles of cADPR and NAADP in pancreatic cells. Acta Biochim Biophys Sin (Shanghai) 44: 719-729, 2012. PMID: 22677461
- Zhou H, Lillis S, Loy RE, Ghassemi F, Rose MR, Norwood F, Mills K, Al-Sarraj S, Lane RJ, Feng L, Matthews E, Sewry CA, Abbs S, Buk S, Hanna M, Treves S, Dirksen RT, Meissner G, Muntoni F, Jungbluth H. Multi-minicore disease and atypical periodic paralysis associated with novel mutations in the skeletal muscle ryanodine receptor (RYR1) gene. Neuromuscul Disord 20: 166-173, 2010. PMID: 20080402
- Zissimopoulos S, Lai FA, and Joachim Krebs Marek M. Ryanodine receptor structure, function and pathophysiology. In: New Comprehensive Biochemistry Elsevier, 2007, p. 287-342.
- Zorzato F, Fujii J, Otsu K, Phillips M, Green NM, Lai FA, Meissner G, MacLennan DH. Molecular cloning of cDNA encoding human and rabbit forms of the Ca2+ release channel (ryanodine receptor) of skeletal muscle sarcoplasmic reticulum. J Biol Chem 265: 2244-2256, 1990. PMID: 2298749
- Zucchi R, Ronca-Testoni S, Di Napoli P, Yu G, Gallina S, Bosco G, Ronca G, Calafiore AM, Mariani M, Barsotti A. Sarcoplasmic reticulum calcium uptake in human myocardium subjected to ischemia and reperfusion during cardiac surgery. J Mol Cell Cardiol 28: 1693-1701, 1996. PMID: 8877779


