Methods Type:
Entry Version:
Citation:
Pancreapedia: Exocrine Pancreas Knowledge Base, DOI: 10.3998/panc.2011.9
| Attachment | Size |
|---|---|
| 277.95 KB |
1. Introduction
Photoaffinity labeling is a powerful technique for the identification of interacting molecules. When it is applied to a ligand-receptor system using a photolabile ligand probe, it provides an opportunity for the identification of the receptor molecule, as well as for the identification of the portion of the receptor that is labeled and even the identification of spatially approximated residues within the docked ligand and receptor. It is particularly useful when being applied to sparse and hydrophobic membrane receptors that are often quite difficult to characterize using other high resolution physicochemical techniques. Photoaffinity labeling has been applied to many pancreatic receptors, including those recognizing secretin (1, 2), cholecystokinin (3, 4), gastrin (5, 6), insulin (7), glucagon (8), glucagon-like peptide-1 (9, 10), glucose-dependent insulinotropic polypeptide (11), and vasoactive intestinal polypeptide (12). These receptors are expressed on every type of cell within the pancreas. In this report, we utilize the photoaffinity labeling of the secretin receptor as an example of the application of this technique.
Secretin is secreted by the S cells in the small intestinal mucosa in response to luminal acid and stimulates biliary and pancreatic ductular bicarbonate and water secretion, thereby regulating the pH of duodenal contents (13). Its receptor is a prototypic member of the B family of G protein-coupled receptors (GPCRs) that includes many important drug targets. Understanding of the molecular basis of ligand binding to this receptor can not only help refine our understanding of its conformation, but also provide insights that may be useful for the rational development of drugs acting at all members within this family of GPCRs.
We have utilized photoaffinity labeling to identify multiple residue-residue approximations between secretin and its receptor, providing extremely useful constraints for building a molecular model of the ligand-bound secretin receptor (1).
In this report, we describe the step-by-step procedures of photoaffinity labeling of the secretin receptor using the [Bpa16,Tyr10]rat secretin-27 probe (1) as an example. This ligand probe incorporates a photolabile moiety, benzoyl phenylalanine (Bpa), in position 16 to replace a serine located within the mid-region of the ligand and a tyrosine in position 10 to replace a naturally-occurring leucine as a site for radioiodination. The steps covered include probe characterization by binding and biological activity assays, probe radioiodination, receptor photoaffinity labeling, peptide mapping of the labeled receptor for identification of candidate fragments and radiochemical sequencing for identification of the specific receptor residue that was covalently labeled.
2. Materials
2.1. Radioiodination
- Iodo-Beads (Pierce Chemical Co.)
- 1 mCi Iodine-125 (Na125I, Perkin Elmer Life Sciences Inc.)
- 15 µg lyophilized [Tyr10]rat secretin-27 (for binding assay) or [Bpa16,Tyr10]rat secretin-27 (for photoaffinity labeling). Both peptides were synthesized in our laboratory.
2.2. Binding assay
- Skatron cell harvester (Skatron Instruments Inc.)
- Glass-fiber filtermats (Skatron Instruments Inc.)
- Iso-Data gamma spectrometer
- Enriched secretin receptor-bearing cell membranes prepared from secretin receptor-bearing CHO-SecR cells using a sucrose gradient centrifugation method (14).
- Krebs-Ringer-HEPES (KRH) binding buffer: 25 mM HEPES, pH 7.4, 104 mM NaCl, 5 mM KCl, 1 mM KH2PO4, 1.2 mM MgSO4, 2 mM CaCl2, 1 mM phenylmethylsulfonyl fluoride, 0.01% soybean inhibitor, and 0.2% bovine serum albumin
- Secretin and photolabile peptide [Bpa16,Tyr10]rat secretin-27 (cold peptide) stock solutions at 1 x 10-4 M
- Radioligand: 125I-[Tyr10]rat secretin-27 (see below for iodination procedure)
2.3. cAMP assay
- Lance cAMP kit (Perkin Elmer Life Sciences Inc.)
- Multichannel pipettor
- KRH cAMP assay buffer: 25 mM HEPES, pH 7.4, 104 mM NaCl, 5 mM KCl, 1 mM KH2PO4, 1.2 mM MgSO4, 2 mM CaCl2, 0.01% soybean trypsin inhibitor, 0.1% bacitracin, 0.2% bovine serum albumin, and 1 mM 3-isobutyl-1-methylxanthine
- Secretin and [Bpa16,Tyr10]rat secretin-27 stock solution at 1 x 10-4 M
- Secretin receptor-bearing CHO-SecR cells: 8,000 cells per well grown in 96-well plates for 48 hours
2.4. Photoaffinity labeling
- Receptor source: secretin receptor-bearing CHO-SecR cell membranes
- KRH photoaffinity labeling buffer: 25 mM HEPES, pH 7.4, 104 mM NaCl, 5 mM KCl, 1 mM KH2PO4, 1.2 mM MgSO4, 2 mM CaCl2, 1 mM phenylmethylsulfonyl fluoride, and 0.01% soybean inhibitor
- Photolabile probe: 125I-[Bpa16,Tyr10]rat secretin-27 (see below for iodination procedure)
- Photolysis apparatus (Rayonet photochemical reactor, Southern New England Ultraviolet, Hamden, CT) equipped with 3500-Å lamps
- Siliconized microcentrifuge tubes
- Siliconized 12 x 75 cm glass tubes. These were prepared by treating glass tubes in 5% AquaSil siliconizing fluid (Pierce Chemical Co.).
2.5. Peptide mapping
- Cyanogen bromide (Pierce Chemical Co.)
- BNPS skatole (Pierce Chemical Co.)
- Endoproteinase Lys-C (Roche Molecular Biochemicals)
- Endoproteinase Asp-N (Roche Molecular Biochemicals)
- 10% NuPAGE gel and MES running buffer (Invitrogen)
- SeeBlue molecular weight markers (Invitrogen)
2.6. Radiochemiscal sequencing
- N-(2-aminoethyl-1)-3-aminopropyl glass beads (Sigma)
- Phenylisothiocyanate (PITC, Pierce Chemical Co.)
- Sulfo-MBS (m-maleimidobenzoyl-N-hydroxysulfosuccinimide ester, Pierce Chemical Co.)
- Purified radiolabeled receptor fragment.
3. Methods
3.1. Radioiodination
- Solubilize an aliquot of 15 µg of [Tyr10]rat secretin-27 (for binding assay) or [Bpa16,Tyr10]rat secretin-27 (for photoaffinity labeling) in 100 µl 20 % (v/v) acetonitrile and 20 % (v/v) methanol.
- Add 100 µl 0.2 M borate buffer, pH 9.0 and 10 µl 1 mCi Na125I.
- Add an Iodo-Bead to the reaction and react for 15 s.
- Transfer the reaction to a new tube and dilute with 0.5 ml 0.1 % trifluoroacetic acid.
- Load the diluted material onto a reversed-phase C18 column and elute the radiolabeled product from the column with a gradient from 100% buffer A consisting of 0.1 % trifluoroacetic acid to 100 % buffer B consisting of 100% acetonitrile over 50 min.
- Collect fractions containing the radioactive peak from the HPLC and store in 100 µl aliquots at –80 ºC.
3.2. Binding assay
- Dilute secretin and [Bpa16,Tyr10]rat secretin-27 to 10-10, 10-9, 10-8, 10-7, 10-6, 10-5 M with KRH binding buffer.
- Dilute CHO-SecR membranes to 0.1 mg protein/ml with KRH binding buffer.
- Dilute radioligand 125I-[Tyr10]rat secretin-27 to 20,000 cpm per 50 µl with KRH binding buffer.
- Label 12 x 75 cm tubes in duplicate: 0, -11, -10, -9, -8, -7, and -6 M concentrations.
- Add 350 µl KRH binding buffer to each tube.
- Add 50 µl diluted peptide: 10-5 to -6 tube, 10-6 to -7 tube, etc. Add 50 µl KRH binding buffer to the control (0) tubes.
- Add 50 µl dilute radioligand to each tube.
- Add 50 µl diluted CHO-SecR membranes to each tube.
- Vortex and incubate at room temperature for 1 h.
- Soak a glass fiber filtermat in 0.3 % polybrene for 1 h.
- Separate bound radioactivity from free radioactivity using a Skatron cell harvester.
- Count filters using an Iso-Data gamma spectrometer.
- Analyze data using GraphPad Prism software package (Figure 1, left panel).
3.3. cAMP assay
- Dilute secretin and [Bpa16,Tyr10]rat secretin-27 from their 10-4 M stock to make 10-12, 10-11, 10-10, 10-9, 10-8, 10-7, and 10-6 M with KRH cAMP assay buffer.
- Check CHO-SecR cells grown in 96-well plate under microscope for cellular confluency, aiming for 80-90 percent.
- Wash cells twice with 100 µL PBS. Use a multichannel pipettor for PBS washes and for perchloric acid, water and KHCO3 additions below.
- Add 50 µl diluted peptide solution to each well (in duplicates), and add 50 µl KRH cAMP assay to the control wells.
- Incubate the plate at 37 °C for 30 min.
- Aspirate peptide solution and add 50 µl of 6 % cold perchloric acid to each well and then shake on an Eppendorf MixMate plate shaker at 1500 rpm for 15 min.
- Add 25 µl cold water and adjust pH of each well to between 5 and 7 with 30 % KHCO3.
- Determine cAMP levels in the cell lysate using the LANCE cAMP kit following the manufacture’s instructions.
- Analyze data using Microsoft Office Exel and GraphPad Prism software package (Figure 1, right panel).

Figure 1. Functional characterization of [Bpa16,Tyr10]rat secretin-27. Left, competition binding curves, reflecting the ability of increasing concentrations of secretin and [Bpa16,Tyr10]rat secretin-27 (Bpa16 probe) to displace the binding of the 125I-[Tyr10]rat secretin-27 radioligand to membranes from CHO-SecR cells. Values represent percentages of the maximal saturable binding that was observed in the absence of competitor and are expressed as means ± S.E.M. of duplicate data from three independent experiments. Right, intracellular cAMP responses in CHO-SecR cells to increasing concentrations of secretin and [Bpa16,Tyr10]rat secretin-27. Values are expressed as means ± S.E.M. of data from three independent experiments performed in duplicate, with data normalized relative to the maximal response to secretin. Reproduced from Journal of Biological Chemistry (1) with permission of the American Society for Biochemistry and Molecular Biology.
3.4. Photoaffinity labeling
- Dilute secretin peptide from 10-4 M stock to make 10-9, 10-8, 10-7, 10-6, and 10-5 M in 1.7 ml siliconized microcentrifuge tubes.
- Label six 1.7 ml siliconized microcentrifuge tubes with 0, -10, -9, -8, -7 and -6 M.
- Add 350 µl KRH photoaffinity labeling buffer to the -10, -9, -8, -7 and -6 tubes and 400 µl to the 0 tube.
- Add 50 µl diluted peptide: 10-5 to -6 tube, 10-6 to -7 tube, etc.
- Add 50 µl CHO-SecR cell membranes (50 µg) to each tube.
- Add 50 µl 125I-[Bpa16-Tyr10]rat secretin-27 (~200,000 cpm) to each tube.
- Vortex to mix each tube and incubate in the dark for 1 h at room temperature.
- Transfer reactions to siliconized 12 x 75 cm glass tubes and photolyze in the photoreactor at 4 °C for 30 min.
- Transfer reactions to siliconized microcentrifuge tubes. Add 500 µl cold KRH photoaffinity labeling buffer and spin at 14,000 rpm for 5 min at 4 ºC.
- Aspirate supernatant. Wash membrane pellets once more with 1 ml cold KRH photoaffinity labeling buffer. Count each tube for record.
- After photolysis, incubate washed membranes in 25 µl 1 x Laemmli SDS sample buffer at room temperature for at least 15 min before loading onto a 10 % mini slab gel. Run the gel at ~150 volts for about 1 h. Use pre-stained molecular weight standards.
- Dry the gel immediately after electrophoresis.
- Expose the gel to an X-ray film in a cassette with an intensifying screen at -80 ºC for various periods of time.
- Develop the film (Figure 2).

Figure 2. Photoaffinity labeling of the secretin receptor with 125I-[Bpa16,Tyr10]rat secretin-27. Shown is a typical autoradiograph of a 10% SDS-PAGE gel used to separate products of affinity labeling of plasma membranes from CHO-SecR cells. The affinity-labeled secretin receptor migrated at approximate Mr = 70,000, and the labeling was inhibited by increasing concentrations of competing unlabeled secretin. Reproduced from Journal of Biological Chemistry (1) with permission.
3.5. Peptide mapping
3.5.1. Receptor purification
- Perform photoaffinity labeling in the absence of competing secretin peptide using 200 µg CHO-SecR membranes and 125I-[Bpa16-Tyr10]rat secretin-27 (~1,000,000 cpm). Set up at least four tubes.
- After photolysis and gel electrophoresis, wrap the wet gel with plastic and expose the gel to an X-ray film in a cassette with intensifying screen at –80 ºC for certain of period of time, dependent on the efficiency of labeling.
- Develop the film, cut labeled receptor bands and crush the gel slices in water in a Dounce homogenizer using the tight setting.
- Transfer gel slurry to a 15-ml tube. Rotate the tube end-over-end for at least 5 h at 4 °C.
- Centrifuge at 2,000 rpm for 5 min. Transfer the supernatant to a 50-ml tube.
- Resuspend the gel slurry with an equal volume of distilled water and pellet by centrifugation. Transfer the supernatant to the 50-ml tube.
- Freeze eluted sample at -80 °C for at least 1 h, and then lyophilize the sample (~14 ml) to dryness.
- Resuspend the lyophilized sample in 300 µl distilled water and equally transfer it to two siliconized microcentrifuge tubes.
- Add 850 µl cold ethanol into each tube and keep at -20 °C for at least 4 h.
- Centrifuge at 14,000 rpm for 30 min at 4 ºC and carefully aspirate the supernatant.
- Resuspend the pellet in 1 ml of cold ethanol and spin at 14,000 rpm for 15 min at 4 ºC.
- Dry the pellet in a SpeedVac for 5 min. Samples are ready for chemical and protease cleavage.
3.5.3. Chemical and protease cleavage
Chemical and/or enzymatic digestion can provide useful information for identification of the covalently labeled portion of a photoaffinity labeled receptor. Cyanogen bromide is the most commonly used chemical method for peptide mapping studies, since it can quantitatively cleave at the carboxyl terminus of all methionine residues within that protein, and since this amino acid is present in limited frequency in most proteins, thereby yielding a limited number of variable size products. BNPS-skatole is another frequently used chemical, which cleaves at the carboxyl terminus of tryptophan residues within a protein. However, the action of this chemical may be limited, and it can miss some possible target sites, making interpretation challenging. Among the many selective proteases possible for peptide mapping, endoproteinase Lys-C and Asp-N are most often used. The former cleaves proteins at the carboxyl terminus of lysine residues, while the latter specifically cleaves the amino-terminal bond of aspartic acid residues and sulfated cysteine residues. The action of proteases is also often limited by access to relevant targets.
3.5.3.1. Cyanogen bromide cleavage
- Dissolve each gel-purified labeled receptor pellet in 60 µl distilled water (if insoluble, add 2 µl 10 % SDS) and add 140 µl 100 % formic acid containing 5 mg cyanogen bromide.
- Flush the reaction with nitrogen and shake in dark for 72 h at room temperature.
- Dry the sample in a SpeedVac. Add 250 µl distilled water to each tube, and dry the sample in a SpeedVac. Repeat this step once.
- Run samples on a 10 % NuPAGE gel in MES-based running buffer. Use SeeBlue molecular weight markers as protein standard for estimation of the apparent molecular size of labeled fragments.
- Expose wet or dried gel to an X-ray film in a cassette with an intensifying screen at –80 ºC for various periods. Use wet gel for autoradiography if bands of interest are needed for purification for sequential cleavage by another chemical or protease.
- Develop the film (Figure 3).
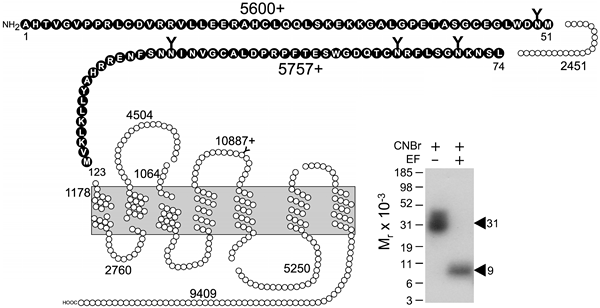
Figure 3. Peptide mapping of the labeled secretin receptor domain by cyanogen bromide (CNBr) cleavage. Shown is a diagram illustrating the theoretical fragments of the secretin receptor resulting from CNBr cleavage. Shown also is a typical autoradiograph of a 10 % NuPAGE gel used to separate the products of CNBr cleavage of the intact secretin receptor affinity labeled with 125I-[Bpa16,Tyr10]rat secretin-27. Cleavage of the labeled secretin receptor yielded a band migrating at approximate Mr = 31,000 and shifting to Mr = 9,000 after deglycosylation by endoproteinase F (EF). The two glycosylated fragments highlighted in heavy circles in the diagram at the receptor amino terminus are the best candidates to represent the domain of labeling. Reproduced from Journal of Biological Chemistry (1) with permission.
3.5.3.2. BNPS-skatole cleavage
- Incubate the gel-purified intact receptor or receptor fragment with 2 mg/ml BNPS-skatole in 70 % acetic acid in a final volume of 200 µl in the dark at 37 °C for 48 h. If cleaving a labeled receptor fragment, cut the fragment band from the NuPAGE gel, elute, lyophilize, precipitate it in 90 % acetone and dry the pellet in a SpeedVac.
- Dry the sample in a SpeedVac. Add 250 µl distilled water to each tube, and dry the sample in a SpeedVac. Repeat this step once.
- Run samples on a 10 % NuPAGE gel in MES-based running buffer. Dry the gel and perform autoradiography.
3.5.3.3. Endoproteinase Lys-C cleavage
- Dissolve ethanol-precipitated intact receptor or receptor fragment in 20 µl endoproteinase Lys-C buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 0.1 % SDS).
- Add 5 µl 0.1 mg/ml endoproteinase Lys-C to the tube and incubate at 37 °C for 18 h.
- Reduce the volume in a SpeedVac and run the sample on a 10 % NuPAGE gel in MES-based running buffer.
- Dry the gel and perform autoradiography.
3.5.3.4. Endoproteinase Asp-N cleavage
- Incubate the ethanol-precipitated intact receptor or receptor fragment with 0.2 µg of endoproteinase Asp-N in 25 µl of digestion buffer containing 50 mM sodium phosphate, pH 8.0, and 0.01 % SDS at 37 °C for 24 h.
- Reduce the volume in a SpeedVac and run the sample on a 10 % NuPAGE gel in MES-based running buffer.
- Dry the gel and perform autoradiography.
3.6. Radiochemical sequencing
- Weigh 10 mg N-(2-aminoethyl-1)-3-aminopropyl glass beads in a 1.5 ml siliconized microcentrifuge tube. The beads, once coupled with the sulfo-MBS bifunctional cross-linking reagent, will be used to immobilize the radiochemically-pure receptor fragment through the thiol groups of its free cysteine residue(s).
- Prepare 10 mM Sulfo-MBS in PBS and add 50 µl 10 mM Sulfo-MBS to the tube containing glass beads. Incubate on ice for 1 h with occasional mixing.
- After incubation, quench amino groups by adding 10 µl 1.0 M Tris-HCl, pH 6.8 and incubate for 10 min on ice.
- Wash twice with 500 µl cold PBS. Aspirate PBS and the beads are ready for coupling the receptor fragment.
- Transfer purified labeled receptor fragment to glass beads as prepared above. Rotate sample at 4 ºC for overnight.
- Wash the beads with 200 µl of methanol for two times, 200 µl of methanol:water (1:1, v:v) for two times, and 200 µl of methanol for two times. Dry beads in a SpeedVac.
- Wash with 50 µl triethylamine for three times. Dry beads in a SpeedVac.
- Add 60 µl triethylamine:methanol (1:7, v:v) for blank cycles or triethylamine:methanol: PITC (1:7:1, v:v:v) for PITC sequencing cycles. Incubate at 50 ºC for 5 min. Before initiating PITC cycle 1, perform at least three cycles of steps excluding PITC to ensure elution of all non-covalently bound radiolabeled fragments.
- Wash beads three times with 400 µl ethyl acetate. Pool washes and count for record. Dry beads in a SpeedVac.
- Incubate beads with 50 µl of trifluoroacetic acid for 5 min at 25 ºC. Wash beads three times with 200 µl methanol. Pool washes and count for record.
- Dry beads in a SpeedVac. Continue next cycle starting from step 1.
- After finishing sequencing, dry all tubes containing methanol washes in a chemical hood. After subtracting the background, calculate the eluted radioactivity in each tube and determine the location of the peak that corresponds with the labeled receptor residue (Figure 4).

Figure 4. Identification of the affinity labeled secretin receptor residue by radiochemical sequencing. Shown is the radioactive elution profile of Edman degradation sequencing of the purified endoproteinase Lys-C fragment (Pro97–Met123) resulting from cleavage of the R96K secretin receptor mutant labeled with 125I-[Bpa16,Tyr10]rat secretin-27. A radioactive peak consistently eluted in cycle 3 that corresponds to covalent labeling of residue Leu99 of the secretin receptor with this probe. Reproduced from Journal of Biological Chemistry (1) with permission.
4. Notes
- Membrane binding assay can be performed using centrifugation or 96-well plate FilterMate Harvester (PerkinElmer). Cell binding assay is also frequently used in our laboratory (9).
- Like most GPCRs, the secretin receptor is a glycosylated protein and deglycosylation is very useful during the process of peptide mapping. This can be done using endoglycosidase F (15) or N-Glycosidase F (Proenzyme) following the procedures described in the manufacture’s manual.
- For some low affinity photoaffinity labeling probes or for low expressing receptor mutants, enrichment using lectin beads (EY Laboratories, Inc.) is very helpful (15).
- Depending on the chemistry of the photolabile residue incorporated in the secretin probe, the wavelength of the UV lamps for photolysis can vary. For benzoyl phenylalanine (Bpa), use 3500-Å lamps. For benzoyl-benzoyl lysine, use 3000-Å lamps.
- When a secretin probe incorporating a photolabile residue in the amino-terminal region of the peptide is used, it is necessary to block the amino terminus of the probe attaching to the receptor fragment for radiochemical sequencing. In this case, use amino group blockers such as acetic anhydride or sulfo-NHS acetate to block both the probe and receptor after photoaffinity labeling but before further chemical and protease cleavage (16). Upon cleavage, new free primary amino group of a receptor fragment will be generated that can be used for radiochemical sequencing Edman degradation. However, it is not necessary to block the ligand probe if the incorporated photolabile residue and the radioiodinated residue are away from the amino terminus, as long as they are not cleaved before the labeled receptor residue to which they covalently attach.
Acknowledgments
This work was supported by grants from the National Institutes of Health (DK46577 and DK32828).
5. References
- Dong M, Lam PC, Pinon DI, Orry A, Sexton PM, Abagyan R, Miller LJ. Secretin occupies a single protomer of the homodimeric secretin receptor complex: insights from photoaffinity labeling studies using dual sites of covalent attachment. J Biol Chem 285:9919-9931, 2010. PMID: 20100828.
- Dong M, Lam PC, Pinon DI, Sexton PM, Abagyan R, Miller LJ. Spatial approximation between secretin residue five and the third extracellular loop of its receptor provides new insight into the molecular basis of natural agonist binding. Mol Pharmacol 74:413-422, 2008. PMID: 18467541.
- Arlander SJ, Dong M, Ding XQ, Pinon DI, Miller LJ. Key differences in molecular complexes of the cholecystokinin receptor with structurally related peptide agonist, partial agonist, and antagonist. Mol Pharmacol 66:545-552, 2004. PMID: 15322246.
- Ding XQ, Dolu V, Hadac EM, Holicky EL, Pinon DI, Lybrand TP, Miller LJ. Refinement of the structure of the ligand-occupied cholecystokinin receptor using a photolabile amino-terminal probe. J Biol Chem 276:4236-4244, 2001. PMID: 11050076.
- Anders J, Bluggel M, Meyer HE, Kuhne R, ter Laak AM, Kojro E, Fahrenholz F. Direct identification of the agonist binding site in the human brain cholecystokininB receptor. Biochemistry 38:6043-6055, 1999. PMID: 10320330.
- Dong M, Liu G, Pinon DI, Miller LJ. Differential docking of high-affinity peptide ligands to type A and B cholecystokinin receptors demonstrated by photoaffinity labeling. Biochemistry 44:6693-6700, 2005. PMID: 15850403.
- Huang K, Chan SJ, Hua QX, Chu YC, Wang RY, Klaproth B, Jia W, Whittaker J, De Meyts P, Nakagawa SH, Steiner DF, Katsoyannis PG, Weiss MA. The A-chain of insulin contacts the insert domain of the insulin receptor. Photo-cross-linking and mutagenesis of a diabetes-related crevice. J Biol Chem 282:35337-35349, 2007. PMID: 17884811.
- Wright DE, Horuk R, Rodbell M. Photoaffinity labeling of the glucagon receptor with a new glucagon analog. Eur J Biochem 141:63-67, 1984. PMID: 6327311.
- Chen Q, Pinon DI, Miller LJ, Dong M. Molecular basis of glucagon-like peptide 1 docking to its intact receptor studied with carboxyl-terminal photolabile probes. J Biol Chem 284:34135-34144, 2009. PMID: 19815559.
- Chen Q, Pinon DI, Miller LJ, Dong M. Spatial approximations between residues 6 and 12 in the amino-terminal region of glucagon-like peptide 1 and its receptor: a region critical for biological activity. J Biol Chem 285:24508-24518, 2010. PMID: 20529866.
- Amiranoff B, Couvineau A, Vauclin-Jacques N, Laburthe M. Gastric inhibitory polypeptide receptor in hamster pancreatic beta cells. Direct cross-linking, solubilization and characterization as a glycoprotein. Eur J Biochem 159:353-358, 1986. PMID: 3530757.
- Ceraudo E, Murail S, Tan YV, Lacapere JJ, Neumann JM, Couvineau A, Laburthe M. The vasoactive intestinal peptide (VIP) alpha-Helix up to C terminus interacts with the N-terminal ectodomain of the human VIP/Pituitary adenylate cyclase-activating peptide 1 receptor: photoaffinity, molecular modeling, and dynamics. Mol Endocrinol 22:147-155, 2008. PMID: 17885205.
- Dong M, Miller LJ. Molecular pharmacology of the secretin receptor. Receptors Channels 8:189-200, 2002. PMID: 12529936.
- Hadac EM, Ghanekar DV, Holicky EL, Pinon DI, Dougherty RW, Miller LJ. Relationship between native and recombinant cholecystokinin receptors: role of differential glycosylation. Pancreas 13:130-139, 1996. PMID: 8829180.
- Dong M, Pinon DI, Asmann YW, Miller LJ. Possible endogenous agonist mechanism for the activation of secretin family G protein-coupled receptors. Mol Pharmacol 70:206-213, 2006. PMID: 16531505.
- Dong M, Li Z, Pinon DI, Lybrand TP, Miller LJ. Spatial approximation between the amino terminus of a peptide agonist and the top of the sixth transmembrane segment of the secretin receptor. J Biol Chem 279:2894-2903, 2004. PMID: 14593094.


