Methods Type:
Entry Version:
Citation:
Pancreapedia: Exocrine Pancreas Knowledge Base, DOI: 10.3998/panc.2011.22
| Attachment | Size |
|---|---|
| 251.09 KB |
The pancreas secretes four types of proteolytic digestive proenzymes, which after activation give rise to trypsin, chymotrypsin, elastase and carboxypeptidase. In humans, three trypsinogen isoforms, cationic trypsinogen (PRSS1), anionic trypsinogen (PRSS2) and mesotrypsinogen (PRSS3); four chymotrypsinogen isoforms, chymotrypsinogen B1 (CTRB1), chymotrypsinogen B2 (CTRB2), chymotrypsinogen C (CTRC), and chymotrypsin-like enzyme-1 precursor (CTRL1) and at least three proelastase isoforms, proelastase 2A (ELA2A), proelastase 3A (ELA3A) and proelastase 3B (ELA3B) are present in the pancreatic juice. Expression of the proelastase 1 (ELA1) gene is evolutionarily suppressed in humans by mutations in the promoter region. The product of the proelastase 2B (ELA2B) gene has not been identified in pancreatic tissue or juice yet and experiments with recombinantly expressed proelastase 2B (ELA2B) indicate that this isoform, if exists, may be catalytically inactive. There are three digestive procarboxypeptidase isoforms expressed in the human pancreas, procarboxypeptidase A1 (CPA1), procarboxypeptidase A2 (CPA2) and procarboxypeptidase B (CPB), also referred to as procarboxypeptidase B1 (CPB1). Here we describe methods we use in our laboratory for the expression, purification, activation and activity measurement of human digestive proteases. Although not tested in a systematic manner, the methodology should be generally applicable to most mammalian pancreatic digestive proteases.
1. Expression and Purification of Recombinant Proteases
1A. Materials
E. coli BL21(DE3) cells (Novagen), LB/agar plates containing 100 µg/mL ampicillin; LB growth medium, ampicillin, isopropyl 1-thio-ß,D-galactopyranoside (IPTG), sucrose, Tris-HCl, K-EDTA, dithiothreitol (DTT), Brilliant Blue R-250 (Coomassie blue) dye; NaCl, KCl, HCl, guanidine-HCl, argon, L-cystine, L-cysteine, L-arginine, reduced glutathione, oxidized glutathione, human embryonic kidney (HEK) 293T cells, D-MEM medium and Opti-MEM reduced serum medium (Invitrogen), fetal bovine serum (FBS), L-glutamine, Lipofectamine 2000 transfection reagent (Invitrogen).
1B. Bacterial Expression and In Vitro Refolding of Recombinant Human Trypsinogens
For high-yield expression, human pancreatic trypsinogens can be produced in E. coli as inclusion bodies (Figure 1). After in vitro refolding, trypsinogen can be purified by ecotin affinity chromatography (1-5). The same protocol applies to cationic trypsinogen (PRSS1), anionic trypsinogen (PRSS2) and mesotrypsinogen (PRSS3).
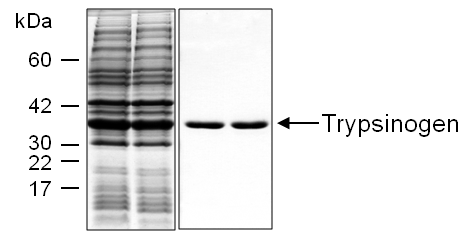
Figure 1. Human cationic trypsinogen (PRSS1) expressed in E. coli (DE3) cells. Left panel: inclusion bodies prepared from 1 mL cell culture. Right panel: ~5 µg purified trypsinogen eluted from the ecotin affinity column. Samples were analyzed by 15% reducing SDS-PAGE followed by Coomassie Blue staining.
- Transform E. coli BL21(DE3) competent cells with pTrap-T7 expression plasmid (available from the authors upon request) containing the appropriate trypsinogen cDNA. Plate the cells onto LB/agar containing 100 µg/mL ampicillin and incubate the plate at 37 oC for 16-18 h.
- For a starter culture, inoculate a streak of transformed cells into 10 mL of LB medium containing 100 µg/mL ampicillin. Incubate the cells overnight at 37 oC with vigorous shaking.
- Next morning, dilute the 10 mL overnight starter culture to 200 mL with fresh LB medium containing 100 µg/mL ampicillin. The OD600 value should be around 0.1. Grow the cell suspension in a 1 liter Erlenmeyer flask with continuous shaking at 37 oC.
- When the OD600 value reaches ~0.5, induce protein expression by adding IPTG to 1 mM final concentration. Grow the culture for an additional 4 h at 37 oC. Confirm protein expression on SDS-PAGE by the following procedure. Remove 1 mL culture; pellet the cells in a microcentrifuge at 16,000 g for 5 min. Resuspend the cell pellet in 0.5 mL Tris-HCl (pH 8.0), 5 mM K-EDTA solution and sonicate the suspension for 20 sec. Collect the inclusion bodies by centrifugation (16,000 g; 5 min, 4 oC in a microcentrifuge) and dissolve the pellet in 100 µL Laemmli sample buffer containing 100 mM DTT. Heat-denature sample at 90 oC for 5 min and run 25 µL on 15% gel. Stain with Coomassie blue.
- Aliquot the cell suspension by 50 mL into Falcon tubes and harvest the cells by centrifugation at 15,000 g for 20 min at 4 oC. Store cell pellets frozen at -20 oC or -80 oC.
- Re-suspend the cell pellet from an 50 mL culture in 6 mL 0.1 M Tris-HCl (pH 8.0), 5 mM K-EDTA solution and aliquot the suspension by 1mL into six microcentrifuge tubes (1.7 mL size).
- Sonicate the cell suspension for 20 sec three times using an ultrasonic cell disruptor.
- Collect the inclusion bodies by centrifuging at 16,000 g for 10 min at 4 oC in a microcentrifuge and resuspend the inclusion body pellets in 0.1 M Tris-HCl (pH 8.0), 5 mM K-EDTA.
- Pellet the inclusion bodies by centrifugation again and repeat the washing two more times. The pellets from the six tubes can be combined to produce a single pellet at the end of the washing procedure. Note that bacterial genomic DNA contamination can make the inclusion body pellet viscous and difficult to pipet. The DNA is removed by the repeated washes and DNAse treatment is usually not necessary.
- After the final wash, resuspend the pellet in 0.5 mL 4 M guanidine-HCl, 0.1 M Tris-HCl (pH 8.0), 2 mM K-EDTA, and add DTT to 30 mM final concentration. We keep DTT at 1 M concentration frozen in small aliquots at -20 oC and freshly thaw an aliquot for each refolding. The guanidine-containing unfolding solution should dissolve the inclusion bodies and the solution should become clear. Incubate the sample(s) for 30 min at 37 oC.
- Remove insoluble material by centrifuging the samples at 16,000 g for 10 min at 4 oC in a microcentrifuge. Save the supernatant.
- Add the supernatant dropwise to 50 mL refolding solution [0.9 M guanidine-HCl, 0.1 M Tris-HCl (pH 8.0), 2 mM K-EDTA, 1 mM L-cystine, 1 mM L-cysteine] while stirring on a magnetic stirrer and continue mixing the solution for 5 min. Transfer the refolding mixture to the refrigerator or cold room and incubate at 4 oC overnight. Next morning, trypsinogens can be purified by ecotin affinity chromatography. Note: To prepare the refolding solution, first add solid L-cystine (oxidized form) to 50 mL refolding buffer in a ~200 mL Erlenmeyer flask and dissolve it by vigorous stirring at room temperature on a magnetic stirrer for 30 min; then add solid L-cysteine (reduced form), which will dissolve quickly. Before adding L-cysteine, equilibrate the refolding solution with argon for a 5 min by blowing the gas into the flask through a pipette tip poked through an aluminum foil cover. Because of the poor solubility of L-cystine, undissolved crystals may remain in the refolding solution. This has no impact on the refolding procedure.
1C. Bacterial Expression and In Vitro Refolding of Recombinant Human Proelastase 2A
We have been unsuccessful so far in expressing human proelastase 2A (ELA2A) in mammalian cell culture. Proelastase 2A (ELA2A)can be expressed as inclusion bodies in E. coli and purified with ecotin-affinity chromatography after refolding, however, yields are lower than those obtained with trypsinogens (6-8). The bacterial expression and inclusion body isolation protocols are identical to those described above for trypsinogens. The unfolding and refolding steps for proelastase 2A (ELA2A) are different. The description below is an updated protocol compared to our previously published method (6).
- Collect the inclusion bodies by centrifugation and add 1 mL of unfolding solution [6 M guanidine-HCl, 50 mM Tris-HCl (pH 8.0) and 5 mM DTT] to ~20 mg of inclusion bodies. Incubate the suspension at room temperature for 2 h.
- Add 90 mL refolding buffer [1.1 M guanidine-HCl, 880 mM L-arginine, 55 mM Tris-HCl (pH 8.2), 21 mM NaCl, 0.88 mM KCl], 1 mL 100 mM K-EDTA, 1 mL 200 mM reduced glutathione, 4 mL 100 mM oxidized glutathione and 2.6 mL distilled water to 5 mL 20 mg/mL unfolded proelastase 2A (ELA2A) solution. Allow refolding to proceed for 16-18 h at 4 oC.
1D. Expression of Human Trypsinogens, Chymotrypsinogens, Proelastases and Procarboxypeptidases in HEK 293T Cells
Expression in of pancreatic digestive protease zymogens can be also achieved in transiently transfected mammalian cell lines (7,8). We routinely use HEK 293T cells. The zymogens are secreted to the conditioned medium, where they typically reach micromolar concentrations. Using this method we were unable to express human proelastase 2A (ELA2A) and we find that expression levels of human cationic trypsinogen (PRSS1) are relatively low (9). On the other hand, human anionic trypsinogen (PRSS2), mesotrypsinogen (PRSS3), chymotrypsinogen B1 (CTRB1), chymotrypsinogen B2 (CTRB2), chymotrypsinogen C (CTRC), chymotrypsin-like enzyme-1 precursor (CTRL1), proelastase 3A (ELA3A), proelastase 3B (ELA3B), procarboxypeptidase A1 (CPA1), procarboxypeptidase A2 (CPA2) and procarboxypeptidase B (CPB) are readily expressed (Figure 2).
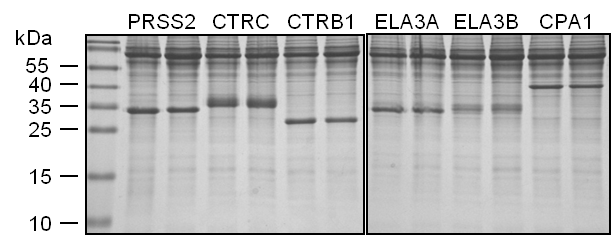
Figure 2. Expression of human pancreatic proteases in HEK 293T cells. Conditioned media (200 µL) were precipitated with 10% trichloroacetic acid (final concentration) and analyzed by 15% reducing SDS-PAGE and Coomassie Blue staining. Experiment performed by Zsolt Rónai in the senior author’s laboratory.
- Grow HEK 293T cells in 20 mL of D-MEM in a 75 cm2 flask with 10% FBS and 4 mM L-glutamine to 70-80% confluence at 37 oC in a tissue culture incubator with a humidified atmosphere containing 5% CO2. We routinely grow 2-3 flasks, but cell mass can be scaled up as needed.
- Add 30 µg of pcDNA3.1 (-) plasmid containing the appropriate proenzyme gene to 2 mL of Opti-MEM (solution A). Add 75 µL of Lipofectamine reagent to 2 mL of Opti-MEM (solution B). After 5 min incubation at room temperature, mix solutions A with B, and incubate the 4 mL transfecting solution for an additional 20 min.
- Remove 4 of the 20 mL D-MEM medium from the cells and replace it with the 4 mL transfecting solution. Incubate for 16-20 h and discard the transfecting solution and wash the cells with 5 mL of Opti-MEM medium.
- Cover the cells with 20 mL of Opti-MEM and harvest the conditioned medium after 48 h of incubation. Replace medium with fresh Opti-MEM and harvest again at 96 h. Only use this second harvest if cell viability is still good, as judged by adherence to the plastic. Pool the 48 h and 96 h harvests.
2. Purification of Pancreatic Serine Proteases
2A. Materials
Liquid chromatography system, e.g. Pharmacia FPLC or a GE Healthcare AKTA Purifier; MonoQ HR 5/5 column (previously Pharmacia, now GE Healthcare), trichloroacetic acid (TCA); LB/agar plate containing 100 µg/mL ampicillin; LB growth medium, ampicillin, quartz cuvette; E. coli BL21(DE3) cells (Novagen), Fernbach flasks, isopropyl 1-thio-ß,D-galactopyranoside (IPTG), sucrose, Tris-HCl, dithiothreitol (DTT), Brilliant Blue R-250 (Coomassie blue) dye; Actigel ALD resin (Sterogene Bioseparations) with coupling solution (1 M sodium cyanoborohydride), Buchner funnel, Na-phosphate, K-EDTA, Parafilm, Adams Nutator or rocking table, freeze-dryer, dialysis tubing (10K MWCO), NaCl, HCl, crystalline bovine trypsin (trypsin TPCK treated; Worthington Biochemical Co), Ni-NTA Superflow Cartridge 5 mL bed volume (Qiagen); imidazole, Vivaspin concentrator (10K MWCO); p-nitrophenyl p'-guanidinobenzoate (PNPGB); dimethyl formamide (DMF).
2B. Purification of Pancreatic Serine Proteases With Ecotin Affinity Chromatography
We routinely purify trypsinogens, chymotrypsinogens and elastases on 2 mL ecotin affinity column using a liquid chromatography system (4). Human chymotrypsinogen B1 (CTRB1) chymotrypsinogen B2 (CTRB2) and proelastase 3A (ELA3A) bind poorly to ecotin but they can be readily purified after activation to chymotrypsins B1 and B2 and elastase 3A. Procarboxypeptdiases or carboxypeptidases do not bind to ecotin. IMPORTANT: because trypsin can activate all other protease zymogens, and chymotrypsin C can process trypsinogen activation peptides to a shorter from, NEVER purify these proenzymes on the same ecotin column. In our laboratory we keep two ecotin columns, one for trypsinogens and one for chymotrypsinogens and proelastases.
- Supplement the 50 mL refolded zymogen solution or the 100-200 mL conditioned media with Tris-HCl (pH 8.0) to 0.1 M concentration and NaCl to 0.2 M concentration. Remove insoluble material by centrifugation at 40,000 g for 20 min at 4 oC. After centrifugation, the zymogen solutions can be also filtered on a 0.22 µM filter; however, this step may result in some protein loss.
- Before use, wash the ecotin column with at least two cycles of washing buffer A [20 mM Tris-HCl (pH 8.0), 0.2 M NaCl] and elution solution B [50 mM HCl]. If the column is clean, no UV peak should be detected when the elution solution B is applied. Equilibrate the column with washing buffer A. Flow rate should be at 2 mL/min.
- Load the refolded zymogen solution or conditioned medium onto the column using a sample loading pump or through pump A at 2 mL/min flow rate.
- Wash the column with washing buffer A until the OD280 returns to the baseline.
- Elute the protease zymogens with the elution solution B at 2 mL/min flow rate and collect the peak. Add Tris-HCl (pH 8.0) to 0.1 M final concentration to chymotrypsinogens and proelastases. Trypsinogens should be stored in elution solution B (50 mM HCl).
- Measure the UV absorption of the purified zymogen solutions at 280 nm in a quartz cuvette and estimate concentrations using the appropriate extinction coefficients. To verify purity, precipitate 50-100 µL of the purified zymogens with trichloroacetic acid (TCA, 10% final concentration), collect the precipitate by centrifugation at 16,000 g for 10 min at 4 oC in a microcentrifuge, dissolve the pellet in 25 µL Laemmli sample buffer containing 100 mM DTT, heat-denature at 90 oC for 5 min, run on a 15% SDS-PAGE gel and stain with Coomassie blue.
Preparation of the Ecotin Affinity Column
Ecotin is a serine-protease inhibitor from the periplasm of E. coli that inhibits all pancreatic serine proteases, including human trypsins, chymotrypsins and elastases. Thus, immobilized ecotin can be utilized as a universal affinity matrix for the purification of pancreatic serine proteases (4). Ecotin also forms relatively tight complexes with the inactive, zymogen forms of pancreatic serine proteases, and can also be used for affinity purification of the proenzymes. Exceptions are chymotrypsinogens B1 (CTRB1) and B2 (CTRB2), which bind poorly to ecotin. In our laboratory, we build our own ecotin affinity chromatography columns, by recombinantly overexpressing ecotin in the periplasmic space of E. coli; isolating the periplasm by osmotic shock and purifying ecotin by trypsin affinity chromatography. Purified ecotin is then immobilized on aldehyde activated resin by reductive amination and loaded into a chromatography column.
Expression of recombinant ecotin in E. coli BL21(DE3)
- Transform E. coli BL21(DE3) cells with pT7-7 plasmid containing the ecotin gene (available from the authors upon request). Spread the cells onto an LB/agar plate containing 100 µg/mL ampicillin and incubate the plate for 16-18 h at 37 oC to select transformants.
- Inoculate a streak of bacteria into 100 mL LB medium with 100 µg/mL ampicillin for an overnight starter culture and grow the cells at 37 oC with vigorous shaking.
- Next morning, dilute 2 × 50 mL of the starter culture to 2×1.2 L with LB medium containing 100 µg/mL ampicillin in two Fernbach flasks (1.2 L each), and grow the cells with continuous shaking at 37 oC until OD600 reaches ~0.5.
- At OD600 ~0.5 induce ecotin expression by adding 1 mM IPTG (final concentration). Grow the culture for an additional 4 h at 37 oC. Confirm ecotin expression levels on SDS-PAGE using the following procedure. Withdraw 1 mL culture and harvest cells by centrifugation in an Eppendorf centrifuge at for 5 min, 4 oC. Resuspend the cell pellet in 0.5 mL osmotic shock buffer [30% sucrose, 20 mM Tris-HCl (pH 8.0), 5 mM K-EDTA] and incubate on ice for 10 min. Pellet the cells by centrifugation (16,000 g; 5 min, 4 oC in a microcentrifuge) and resuspend the pellet in 100 µL cold water. Incubate on ice for 5 min and centrifuge the cells at 16,000 g; for 10 min, at 4 oC in a microcentrifuge. Add 100 µL Laemmli sample buffer containing 100 mM DTT. Heat-denature sample at 90 oC for 5 min and run 25 µL on a 15% gel. Stain with Coomassie blue.
- Harvest the cells by centrifugation at 15,000 g for 20 min at 4 oC.
Preparation of periplasmic fraction
- For best results, periplasm should be isolated immediately after growth. Alternatively, cells may be kept on ice overnight in pellet form, but this typically results in some cell lysis which contaminates the periplasm.
- Resuspend the cell pellet in 400 mL of 30% sucrose, 20 mM Tris-HCl (pH 8.0), 5 mM K-EDTA. Incubate the suspension on ice for 10 min.
- Pellet the cells by centrifugation at 15,000 g for 15 min.
- Resuspend the cells in 200 mL ice cold water, incubate the sample on ice for 10 min.
- Spin down the cells at 40,000 g for 15 min. The supernatant, containing the periplasmic fraction, is removed and saved. A second centrifugation step may be necessary to completely clarify the supernatant.
- Add 5 mL 1 M Tris-HCl (pH 8.0) and 10 mL 5 M NaCl to the supernatant. The procedure may be paused here and the periplasm stored overnight at 4 oC.
Preparation of trypsin affinity column for ecotin purification
- Resuspend the Actigel ALD resin (supplied in 20% ethanol) by turning the bottle upside-down a few times. Insert a 60 mL Buchner funnel into a side-arm flask through a rubber stopper and attach the side arm to the house vacuum through a hose. Pour approximately 20 mL resin into the funnel and wash several times with 50 mM Na-phosphate (pH 7.5) or with PBS (pH 7.4) under suction to remove the ethanol and to equilibrate with the coupling buffer. The coupling buffer must be free of amines, therefore, phosphate-based buffers are recommended. Do not use Tris buffer or other amine-containing buffers.
- Disconnect the vacuum and resuspend the resin in the funnel in ~5 mL phosphate buffer or PBS. Transfer the resuspended resin from the funnel to a 50 mL Falcon tube and allow to settle.
- Remove the supernatant from the settled resin with a pipette. Add 14 mL phosphate buffer or PBS and 80 mg crystalline bovine trypsin to the remaining ~16 mL wet resin. This corresponds to about 5 mg protein per mL resin immobilization ratio. Finally, add 3.4 mL 1 M coupling solution (sodium cyanoborohydride) to 0.1 M final concentration.
- Seal the Falcon tube’s cap with Parafilm and incubate the tube at room temperature for 1-3 h with gentle rocking on an Adams Nutator. You may also move the reaction to the 4oC cold room and leave overnight with gentle rocking on an Adams Nutator.
- Pour the resin into a ~8 mL volume empty chromatography column and remove unbound trypsin and contaminants by washing the column two or three times, alternating between 20 mM Tris-HCl (pH 8.0), 0.2 M NaCl buffer and 50 mM HCl solution. Store the column equilibrated in 20 mM Tris-HCl (pH 8.0), 0.2 M NaCl in the refrigerator at 4oC.
Purification of ecotin by trypsin-affinity chromatography
- Centrifuge the ~220-250 mL supernatant for 15 min at 30,000 g at 4 oC and load on the trypsin affinity column in five runs of 50 mL each. Collect the flow-through from all runs and pool. Re-apply the pooled flow-through to the column.
- Wash the column with 20 mM Tris-HCl (pH 8.0), 0.2 M NaCl. Elute the ecotin with 50 mM HCl. Pool the eluates from all runs. Estimate the concentration and yield of ecotin by reading the absorbance at 280 nm using a calculated molar extinction coefficient of 23,045 M-1 cm-1 and the monomeric molecular mass of 16,099.5 Da. Typical yields vary between 20 and 40 mg of purified ecotin.
- Dialyze the eluate against two changes of 3.5 L distilled water or 1 mM HCl overnight. Confirm ecotin concentration and yield again, as described above. Purity may be verified by subjecting samples of 1-5 µL to SDS-PAGE. There may be low levels of ecotin degradation products present which are presumably generated by tryptic cleavages during chromatography on the trypsin column. Although not necessary, you may obtain a precise concentration of functional ecotin in the preparation by titrating against active-site titrated bovine trypsin.
- Lyophilize the dialyzed ecotin. Aliquot the dialyzed sample to 50 mL Falcon tubes by ~15 mL each. Freeze the ecotin solution onto the tube walls by immersing and slowly rotating each tube in a Dewar flask with liquid nitrogen. Cover the tubes with Parafilm and punch holes in the Parafilm to let water evaporate. Place tubes in a larger freeze-dry bottle or flask and lyophilize overnight. Store the lyophilized ecotin at -20 oC until use.
Preparation of immobilized ecotin and the ecotin column
About 30 mg ecotin should be used for column preparation. If needed, ecotin from two separate purifications should be combined.
- Dissolve the lyophilized ecotin in 3 mL 50 mM Na-phosphate (pH 7.5) buffer or PBS (pH 7.4) to 10 mg/mL final concentration.
- Resuspend the Actigel ALD resin (supplied in a plastic bottle in 20% ethanol solution) by turning the bottle upside-down a few times. Insert a 60 mL Buchner funnel into a side-arm flask through a rubber stopper and attach the side arm to the house vacuum through a hose. Pour approximately 10 mL resin into the funnel and wash several times with 50 mM Na-phosphate (pH 7.5) or with PBS (pH 7.4) under suction to remove the ethanol and to equilibrate with the coupling buffer. The coupling buffer must be free of amines, therefore, phosphate-based buffers are recommended. Do not use Tris buffer or other amine-containing buffers.
- Disconnect the vacuum and resuspend the resin in the funnel in ~5 mL phosphate buffer or PBS. Transfer the resuspended resin from the funnel to a 15 mL Falcon tube and allow to settle (~7 mL packed volume). Remove the supernatant with a pipette.
- Add ~3 mL ecotin solution to the resin, gently resuspend by turning the tube upside-down several times and add 1.2 mL 1 M coupling solution (sodium cyanoborohydride) to 0.1 M final concentration, and incubate the reaction at room temperature on an Adams Nutator for 3 h.
Load the ecotin resin into an ~2 mL chromatography column and wash the column with 50 mM HCl to remove unbound ecotin and contaminants and store the column equilibrated with 20 mM Tris-HCl (pH 8.0), 0.2 M NaCl buffer at 4 oC. The remaining immobilized ecotin can be stored at 4 oC for months and used to make new ecotin columns as needed.
2C. Purification of human procarboxypeptidases A1, A2 and B1 with MonoQ ion-exchange chromatography
Procarboxypeptidases or their active forms do not bind to ecotin. Purification of these proenzymes from conditioned media can be achieved using conventional ion-exchange chromatography (8).
- Dialyze conditioned media containing procarboxypeptidase against 20 mM Tris-HCl (pH 8.0) and load directly onto a MonoQ HR 5/5 column at 1 mL/min flow rate through one of the pumps of a Pharmacia FPLC system or using a loading pump of the AKTA system.
- Develop the column with a 0-0.5 M NaCl gradient applied over 30 min at a flow rate of 1 mL/min.
- Collect 1 mL fractions and analyze peak fraction by running 15 µL on a 15% SDS-PAGE gel and stain with Coomassie Blue.
- Use only the purest one or two fractions for experiments. Estimate concentrations of the purified zymogen solutions from their ultraviolet absorbance at 280 nm, using extinction coefficients calculated with the web-based tool at http://ca.expasy.org/tools/protparam.html. This method usually yields preparations of high but incomplete purity (80-95%). An affinity tag based method (e.g. see purification of His-tagged zymogens on a nickel column) is recommended if the presence of the affinity tag is not a concern.
2D. Purification of His-tagged pancreatic proenzymes with Ni-affinity chromatography
The use of C-terminal affinity tags can significantly facilitate purification of recombinantly expressed pancreatic proenzymes (7). We routinely purify chymotrypsinogen C (CTRC), chymotrypsinogen B1 (CTRB1), chymotrypsinogen B2 (CTRB2) carrying an affinity tag composed of 10 histidine residues (10His tag). We also tested the method with success for His-tagged proelastase 2A (ELA2A), proelastase 3A (ELA3A) and proelastase 3B (ELA3B) as well as procarboxypeptidase A1 (CPA1). The use of a 6His tag should be as good as longer tags. Note that purification of active pancreatic proteases using this method is less consistent, probably because of partial proteolytic removal of the affinity tags.
- Wash an Ni-NTA Superflow Cartridge (Qiagen) with successive changes of NPI-20 buffer [50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, 20 mM imidazole], NPI-250 buffer [50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, 250 mM imidazole] and NPI-20 solutions at 4 mL/min flow rate.
- Load the conditioned medium onto the cartridge at 4 mL/min flow rate through one of the pumps of a Pharmacia FPLC system or using a loading pump on the AKTA system.
- Wash the cartridge with NPI-20 buffer until the OD280 returns to baseline.
- Elute the protein with NPI-250 buffer at 2 mL/min flow rate and collect 5 mL fractions.
- Precipitate 100 µL of the fractions by 10% trichloroacetic acid (TCA, final concentration), collect the precipitate by centrifugation at 16,000 g for 10 min at 4 oC in a microcentrifuge, resuspend the pellet in 25 µL Laemmli sample buffer containing 100 mM DTT, heat denature at 90 oC for 5 min, run on a 15% SDS-PAGE gel and stain with Coomassie blue.
- Pool peak fractions and dialyze overnight against two changes of 3.5 liters of 50 mM NaH2PO4 (pH 8.0) buffer containing 300 mM NaCl.
- Concentrate the dialyzed proenzyme solution using a Vivaspin concentrator, as needed.
3. Concentration Determination by Active Site Titration
For most experiments, concentration estimated from UV absorbance is acceptable, as long as consistently applied. If precise concentrations of functionally active proteases are required, active site titration with ecotin can be performed (7). Dissolve crystalline bovine trypsin in 0.1 M Tris-HCl (pH 8.0), 1 mM CaCl2 at an approximately 50 µM concentration and active-site titrate using p-nitrophenyl p'-guanidinobenzoate (PNPGB). Prepare a 10 mM PNPGB stock solution in dimethyl formamide (DMF). To 990 µL trypsin solution add 10 µL 10 mM PNPGB, mix quickly and thoroughly, and measure OD immediately at 410 nm. Molarity of active trypsin present is calculated from the formula OD×6.025×10-5. Next, determine the concentration of a purified ecotin solution by titration against the active-site titrated bovine trypsin. This titrated ecotin batch will serve then as a universal titrant for all human pancreatic serine proteases. Perform titrations using protease concentrations at least two orders of magnitude above the equilibrium dissociation constants (KD) of ecotin with human pancreatic proteases. This may be impractical with the relatively weakly binding elastase 3A (ELA3A) which can be titrated at a concentration ~5-10-times above KD.
4. Measurment of Protease Activity with Peptide Subtrates
The activity of proteases can be determined using chromogenic or fluorogenic peptide substrates. These substrates are equally useful for assaying purified proteases or protease activity in conditioned medium, pancreatic juice or pancreatic tissue homogenate. We prefer chromogenic substrates with a p-nitroaniline (pNA) leaving group, because the initial substrate concentrations can be more easily defined and the substrate consumption can be readily converted to concentrations using the extinction coefficient of pNA, which is 10,400 M-1·cm-1 at 405 nm. Please note that in microplate readers the extinction coefficient needs to be corrected for the shorter light path. We routinely use N-CBZ-Gly-Pro-Arg-pNA for trypsin, N-Suc-Ala-Ala-Pro-Phe-pNA for chymotrypsin, N-Suc-Ala-Ala-Pro-Ala-pNA for ELA3A and ELA3B and N-Suc-Ala-Ala-Pro-Leu-pNA for ELA2A. Release of the yellow pNA is followed for 1-2 min at 405 nm in 0.1 M Tris-HCl, (pH 8.0), 1 mM CaCl2, and 0.05% Tween 20, at 22 °C, in a Spectramax Plus 384 microplate reader (Molecular Devices). The microplate format also allows for easy assay of duplicate or triplicate samples, as needed. Initial velocity of the enzymatic reaction is determined from the linear portion of curve and expressed in mOD·min-1 units which can be then converted to pNA concentration liberated per time unit (e.g. nM·pNA·sec-1) (Figure 3). Note that above 500 mOD·min-1 the initial rate determination becomes unreliable and measurements need to be repeated with more dilute enzyme concentrations.
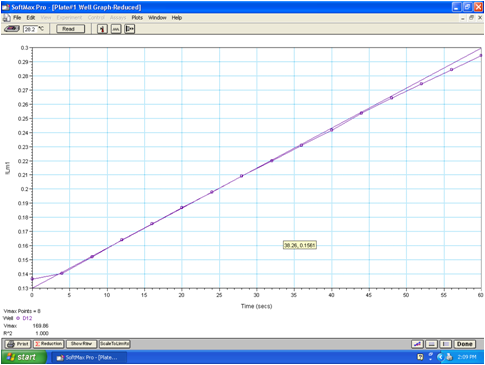
Figure 3. Protease activity measurement. The captured screenshot shows the 16 measurements the plate reader took at 405 nm wavelength during a 1 min time course. The rate of substrate hydrolysis was determined by a linear fit to the initial 8 points of the curve. The rate, erroneously denoted as Vmax by the software, is expressed in mOD/min units.
4A. Materials
Microplate reader, e.g. Spectramax Plus 384 (Molecular Devices), Tris-HCl, CaCl2, Tween 20, dimethylformamide (DMF), N-CBZ-Gly-Pro-Arg-pNA (Bachem L-1560 or Sigma-Aldrich C2276), N-Suc-Ala-Ala-Pro-Phe-pNA (Bachem L-1400), N-Suc-Ala-Ala-Pro-Ala-pNA (Bachem L-1775), N-Suc-Ala-Ala-Pro-Leu-pNA (Bachem L-1390) or N-Glt-Ala-Ala-Pro-Leu-pNA (Peptides International SGL-3129), N-[4-methoxyphenylazoformyl]-L-phenylalanine (Bachem M2245), N-[4-methoxyphenylazoformyl)-L-arginine (Bachem M2525), human enteropeptidase (R&D Systems).
4B. Measurement of trypsin, chymotrypsin and elastase activity on purified enzymes
Prepare a 0.2 mM substrate stock solution by dissolving the solid peptide substrate in 0.25% volume of dimethylformamide (DMF) and adding 99.75% volume of assay buffer [0.1 M Tris (pH 8.0), 1 mM CaCl2 and 0.05% Tween 20]. E.g. for 200 mL of trypsin substrate, dissolve 25 mg N-CBZ-Gly-Pro-Arg-pNA in 0.5 mL DMF and dilute to 200 mL with buffer.
Add 1-5 µL enzyme solution to 50 µL final volume of assay buffer (e.g. 2 µL enzyme to 48 µL assay buffer) and start the reaction by adding 150 µL 0.2 mM substrate solution.
4C. Measurement of carboxypeptidase activity on purified enzymes
CPA1 and CPA2 activity can be determined using the N-[4-methoxyphenylazoformyl]-L-phenylalanine substrate at 60 µM final concentration (8). The assay mix contains 4 µL CPA1/CPA2 enzyme (80 nM final concentration), 86 µL assay buffer (0.1 M Tris-HCl (pH 8.0), 1 mM CaCl2 and 0.05% Tween 20) and 10 µL 0.6 mM substrate dissolved in assay buffer. Start the assay by adding the substrate. CPB1 activity can be measured with N-[4-methoxyphenylazoformyl)-L-arginine under the same conditions (8). The decrease in absorbance is followed at 350 nm for 2 min in a SPECTRAmax Plus 384 microplate reader. Rates of substrate consumption obtained in mOD·min-1 units can be converted to nM·s-1 values using an extinction coefficient of 19,000 M-1·cm-1 corrected for the shorter path length of the microplate wells. Initial rate determinations become unreliable above 200 mOD·min-1 and measurements need to be repeated with more dilute enzyme concentrations. Note that addition of ZnCl2 to the assay is not necessary.
4D. Measurement of protease activity from conditioned media
Measurement of trypsin activity from conditioned media
Supplement 5-20 µL conditioned medium from HEK 293T cells to 50 µL volume with 0.1 M Tris-HCl (pH 8.0), 1 mM CaCl2 (final concentrations) and activate trypsinogen by incubating with 1 µL 1.4 µg/mL human enteropeptidase (R&D Systems) for 1 h at 37 oC. Measure trypsin activity by adding 150 µL of N-CBZ-Gly-Pro-Arg-p-nitroanilide to 0.14 mM final concentration.
Measurement of chymotrypsin activity from conditioned media
To activate chymotrypsinogens, add 5 µL 1 M Tris-HCl (pH 8.0), 5 µL 0.1 M CaCl2 and 2.5 µL 2 µM human cationic trypsin (100 nM final concentration) to 37.5 µL conditioned medium from HEK 293T cells and incubate for 1 h at 37 oC. Measure chymotrypsin activity by adding 150 µL of N-Suc-Ala-Ala-Pro-Phe-p-nitroanilide to 0.15 mM final concentration.
Measurement of elastase activity from conditioned media
To activate proelastases, add 5 µL 1 M Tris-HCl (pH 8.0), 5 µL 10 mM CaCl2 and 2.5 µL 2 µM human cationic trypsin (100 nM final concentration) to 37.5 µL conditioned medium from HEK 293T cells and incubate for 1 h at 37 oC. Measure elastase activity by adding 150 µL of N-Suc-Ala-Ala-Pro-Ala-p-nitroanilide to 0.15 mM final concentration.
Measurement of carboxypeptidase activity from conditioned media
To activate procarboxypeptidases, add 5 µL 1 M Tris-HCl (pH 8.0), 5 µL 10 mM CaCl2, 2 µL 2 µM human cationic trypsin (100 nM final concentration), 2 µL 2 µM human chymotrypsin C (100 nM final concentration) and 6 µL water to 20 µL conditioned medium from HEK 293T cells and incubate for 1 h at 37 oC. Add 50 µL assay buffer [0.1 M Tris-HCl (pH 8.0), 1 mM CaCl2 and 0.05% Tween 20) and measure carboxypeptidase activity by adding 10 µL of 0.6 mM N-[4-methoxyphenylazoformyl]-L-phenylalanine (for CPA1 and CPA2) or 10 µL of 0.6 mM N-[4-methoxyphenylazoformyl)-L-arginine (for CPB1).
Acknowledgement
Work in the senior author's laboratory was supported by NIH grants R01 DK058088, R01 DK082412 and R01 DK082412-S2 (ARRA).
5. References
- Sahin-Tóth M. (2000) Human cationic trypsinogen. Role of Asn-21 in zymogen activation and implications in hereditary pancreatitis. J Biol Chem 275, 22750-22755. PMID: 10801865
- Sahin-Tóth M. Tóth M. (2000) Gain-of-function mutations associated with hereditary pancreatitis enhance autoactivation of human cationic trypsinogen. Biochem Biophys Res Commun 278, 286-289. PMID: 11097832
- Király, O., Guan, L., Szepessy, E., Tóth, M., Kukor, Z., Sahin-Tóth, M. (2006) Expression of human cationic trypsinogen with an authentic N terminus using intein-mediated splicing in aminopeptidase P deficient Escherichia coli. Protein Expr Purif 48, 104-111. PMID: 16542853
- Lengyel Z, Pál G, Sahin-Tóth M. (1998) Affinity purification of recombinant trypsinogen using immobilized ecotin. Protein Expr Purif 12, 291-294. PMID: 9518472
- Király O, Guan L, Sahin-Tóth M. (0211) Expression of recombinant proteins with uniform N-termini. Methods Mol Biol 705, 175-194. PMID: 21125386
- Szepessy E, Sahin-Tóth M. (2006) Inactivity of recombinant ELA2B provides a new example of evolutionary elastase silencing in humans. Pancreatology 6, 117-122. PMID: 16327289
- Szabó A, Héja D, Szakács D, Zboray K, Kékesi KA, Radisky ES, Sahin-Tóth M, Pál G. (2011) High affinity small protein inhibitors of human chymotrypsin C (CTRC) selected by phage display reveal unusual preference for P4' acidic residues. J Biol Chem 286, 22535-22545. PMID: 21515688
- Szmola R, Bence M, Carpentieri A, Szabó A, Costello CE, Samuelson J, Sahin-Tóth M. (2011) Chymotrypsin C is a co-activator of human pancreatic procarboxypeptidases A1 and A2. J Biol Chem 286, 1819-1827. PMID: 21098023
- Kereszturi E, Sahin-Tóth M. (2009) Intracellular autoactivation of human cationic trypsinogen mutants causes reduced trypsinogen secretion and acinar cell death. J Biol Chem 284, 33392-33399. PMID: 19801634


