Entry Version:
Citation:
Pancreapedia: Exocrine Pancreas Knowledge Base, DOI: 10.3998/panc.2012.9
| Attachment | Size |
|---|---|
| 203.62 KB |
Protein Symbols: CUX1, CUTL1 (cut-like homeobox 1)
Gene Symbol: CUX1
1. General function
The family of CUX/CDP-proteins is a group of transcription factors that are highly conserved among metazoans. They contain one homeodomain and at least one ‘CUT-repeat’.
Early studies suggested that the expression of CUX1 in mammalians is restricted to proliferating and undifferentiated cells (20, 27, 28). However, more recent publications demonstrated that CUX1 expression can also be found in terminally differentiated cells of several tissues (5, 10). Proteolytic processing of the full length protein (p200) by Cathepsin L and other not yet identified proteases generates several isoforms of CUX1 (p150, p110, p90 and p80) that lack one or two of the three N-terminal CUT-repeats (7, 8, 15, 25). Another shortened isoform (p75) is generated by the usage of an alternative transcriptional start site (8, 23).
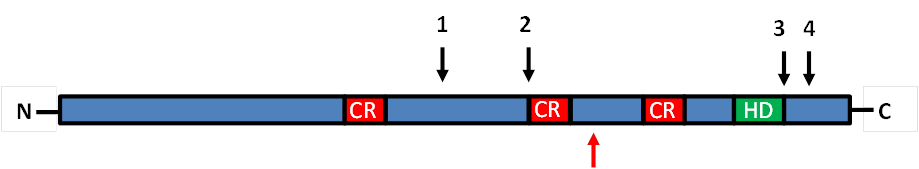
Fig.1. Domain structure of human full length-CUX1 (p200). Cut-Repeats are shown in red and the homeodomain in green. Black arrows indicate protease cleavage sites that lead to generation of the different CUX1 isoforms (p150 – 3; p110 – 1; p90 – 2; p80 – 1,4) . The usage of an alternative transcriptional start site generates the shortened isoform p75 (red arrow).
The presence of DNA binding cut-repeats in the CUX1 proteins influences their interaction with DNA and their transcriptional activity. In most circumstances, the function of CUX1 has been described as transcriptional repressor that represses gene expression by at least three different mechanisms: displacement of transcriptional activators, recruitment of histone deacetylases or recruitment of histone lysine methyltransferases (1, 3, 4, 11, 13, 14, 19, 26). Numerous recent reports suggest that CUX1 functions also as transcriptional activator. However, only little is known about the underlying mechanisms.
Three main cellular processes have been described to be influenced by CUX1: cell proliferation, cell motility/invasiveness and apoptosis. The pro-proliferative effects are due to a shortened G1-phase mainly mediated by the p110 CUX1 isoform (6, 18, 24).
Several studies showed that in vitro knockdown of CUX1 decreases cell migration and invasion in different human cell lines whereas stable overexpression of p75 and p110 CUX1 increases cell motility and invasiveness (2, 9, 17). In tail vein injection experiments CUX1-shRNA expressing cells revealed reduced pulmonary colony formation, whereas CUX1 stably overexpressing cells led to an increased number of lung metastases (2, 17).
The anti-apoptotic effects of CUX1 have been shown in cancer cells in vitro and in xenograft mouse models (21, 22).
2. CUX1 in the Pancreas
CUX1 was found to be significantly overexpressed in human pancreatic cancer tissues compared to normal pancreas by in situ hybridization and immunohistochemistry (21). Furthermore the CUX1 expression level increases during cancer progression as high-grade tumors show higher CUX1 levels than low-grade tumors (17). One possible explanation for this observation is the increasing concentration of TGFbeta during pancreatic tumorigenesis as TGFbeta treatment stimulates expression of CUX1 mRNA and protein levels in several cell types including pancreatic cancer cells (17). Another known stimulator of CUX1 expression in pancreatic cancer cells is IGF1 that induces CUX1 expression via phosphatidylinositol 3-kinase (21).
Studies have shown that CUX1 has pro-invasive, pro-proliferative and anti-apoptotic effects in pancreatic cancer cells in vitro and in xenograft mouse models (16, 21, 22). SiRNA mediated knockdown of CUX1 increases TNFalpha- and TRAIL-induced cell death whereas overexpression of CUX1 rescues from apoptosis. Additionally, treatment of xenograft tumours with siRNA for CUX1 lead to retarded tumour growth and increased apoptosis (21, 22). Mediators of these effects are, at least in part, the CUX1 downstream targets WNT5A and the glutamate receptor GRIA3 (21, 22).
3. Tools for studies of CUX1
a. Antibodies
Rabbit polyclonal and mouse monoclonal antibodies have been raised against CUX1. Santa Cruz sells several goat, rabbit and mouse antibodies against CUX1. We used SC-13024 for WB and Immunoprecipitation but did not test it for other applications.
Several Anti-CUX1 antibodies are also available from Abcam. For IHC we use the mouse monoclonal antibody ab54583.
b. Mouse lines
Cadieux et al. created FVB mice transgenic for p75 and p110 CUX1 downstream of the mammary tumor virus (MMTV)-long terminal repeat that leads to expression specifically in mammary epithelial cells (2).
Ledford et al. generated a C57Bl/6 x C3H mouse expressing murine CUX1 under the control of the CMV promotor (12).
4. References
- Ai W, Toussaint E, Roman A. CCAAT displacement protein binds to and negatively regulates human papillomavirus type 6 E6, E7, and E1 promoters. J.Virol., 73: 4220-4229, 1999. PMID 10196318
- Cadieux C, Kedinger V, Yao L, Vadnais C, Drossos M, Paquet M, Nepveu A. Mouse mammary tumor virus p75 and p110 CUX1 transgenic mice develop mammary tumors of various histologic types. Cancer Res., 69: 7188-7197, 2009. PMID 19738070
- Catt D, Hawkins S, Roman A, Luo W, Skalnik DG. Overexpression of CCAAT displacement protein represses the promiscuously active proximal gp91(phox) promoter. Blood, 94: 3151-3160, 1999. PMID 10556202
- Catt D, Luo W, Skalnik DG. DNA-binding properties of CCAAT displacement protein cut repeats. Cell MolBio (Noisy.-le-grand)., 45: 1149-1160, 1999. PMID 10643964
- Ellis T, Gambardella L, Horcher M, Tschanz S, Capol J, Bertram P, Jochum W, Barrandon Y, Busslinger M. The transcriptional repressor CDP (Cutl1) is essential for epithelial cell differentiation of the lung and the hair follicle. Genes Dev., 15: 2307-2319, 2001. PMID 11544187
- Goulet B, Baruch A, Moon NS, Poirier M, Sansregret LL, Erickson A, Bogyo M, Nepveu A. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol Cell., 14: 207-219, 2004. PMID 15099520
- Goulet B, Truscott M, Nepveu A. A novel proteolytically processed CDP/Cux isoform of 90 kDa is generated by cathepsin L. Biol Chem., 387: 1285-1293, 2006. PMID 16972798
- Goulet B, Watson P, Poirier M, Leduy L, Bérubé G, Meterissian S, Jolicoeur P, Nepveu A. Characterization of a tissue-specific CDP/Cux isoform, p75, activated in breast tumor cells. Cancer Res., 62: 6625-6633, 2002. PMID 12438259
- Kedinger V, Sansregret L, Harada R, Vadnais C, Cadieux C, Fathers K, Park M, Nepveu A. p110 CUX1 homeodomain protein stimulates cell migration and invasion in part through a regulatory cascade culminating in the repression of E-cadherin and occludin. J Biol Chem., 284: 27701-27711, 2009. PMID 19635798
- Khanna-Gupta A, Zibello T, Sun H, Lekstrom-Himes J, Berliner N. C/EBP epsilon mediates myeloid differentiation and is regulated by the CCAAT displacement protein (CDP/cut). Proc Natl Acad Sci USA., 98: 8000-8005, 2001. PMID 11438745
- Kim EC, Lau JS, Rawlings S, Lee AS. Positive and negative regulation of the human thymidine kinase promoter mediated by CCAAT binding transcription factors NF-Y/CBF, dbpA, and CDP/cut. Cell Growth Differ., 8: 1329-1338, 1997. PMID 9419421
- Ledford AW, Brantley JG, Kemeny G, Foreman TL, Quaggin SE, Igarashi P, Oberhaus SM, Rodova M, Calvet JP, Vanden Heuvel GB. Deregulated expression of the homeobox gene Cux-1 in transgenic mice results in downregulation of p27(kip1) expression during nephrogenesis, glomerular abnormalities, and multiorgan hyperplasia. Dev Biol., 245: 157-171, 2002. PMID 11969263
- Li S, Moy L, Pittman N, Shue G, Aufiero B, Neufeld EJ, LeLeiko NS, Walsh MJ. Transcriptional repression of the cystic fibrosis transmembrane conductance regulator gene, mediated by CCAAT displacement protein/cut homolog, is associated with histone deacetylation. J Biol.Chem., 274: 7803-7815, 1999. PMID 10075672
- Lievens PM, Donady JJ, Tufarelli C, Neufeld EJ. Repressor activity of CCAAT displacement protein in HL-60 myeloid leukemia cells. J Biol Chem., 270: 12745-12750, 1995. PMID 7759529
- Maitra U, Seo J, Lozano MM, Dudley JP. Differentiation-induced cleavage of Cutl1/CDP generates a novel dominant-negative isoform that regulates mammary gene expression. Mol Cell Biol., 26: 7466-7478, 2006. PMID 17015474
- Michl P, Downward J. CUTL1: a key mediator of TGFbeta-induced tumor invasion. Cell Cycle, 5: 132-134, 2006. PMID 16357536
- Michl P, Ramjaun AR, Pardo OE, Warne PH, Wagner M, Poulsom R, D'Arrigo C, Ryder K, Menke A, Gress T, Downward J. CUTL1 is a target of TGF(beta) signaling that enhances cancer cell motility and invasiveness. Cancer Cell, 7: 521-532, 2005. PMID 15950902
- Moon NS, Premdas P, Truscott M, Leduy L, Berube G, Nepveu A. S phase-specific proteolytic cleavage is required to activate stable DNA binding by the CDP/Cut homeodomain protein. Mol Cell Biol., 21: 6332-6345, 2001. PMID 11509674
- Nishio H, Walsh, MJ. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc Natl Acad Sci USA., 101: 11257-11262, 2004. PMID 15269344
- Pattison S, Skalnik DG, Roman A. CCAAT displacement protein, a regulator of differentiation-specific gene expression, binds a negative regulatory element within the 5' end of the human papillomavirus type 6 long control region. J Virol., 71: 2013-2022, 1997. PMID 9032333
- Ripka S, Neesse A, Riedel J, Bug E, Aigner A, Poulsom R, Fulda S, Neoptolemos J, Greenhalf W, Barth P, Gress TM, Michl P. CUX1: target of Akt signalling and mediator of resistance to apoptosis in pancreatic cancer. Gut, 59: 1101-1110, 2010. PMID 20442202
- Ripka S, Riedel J, Neesse A, Griesmann H, Buchholz M, Ellenrieder V, Moeller F, Barth P, Gress TM, Michl P. Glutamate receptor GRIA3--target of CUX1 and mediator of tumor progression in pancreatic cancer. Neoplasia., 12: 659-667, 2010. PMID 20689760
- Rong Zeng W, Soucie E, Sung Moon N, Martin-Soudant N, Bérubé G, Leduy L, Nepveu A. Exon/intron structure and alternative transcripts of the CUTL1 gene. Gene, 241: 75-85, 2000. PMID 10607901
- Sansregret L, Goulet B, Harada R, Wilson B, Leduy L, Bertoglio J, Nepveu A. The p110 isoform of the CDP/Cux transcription factor accelerates entry into S phase. Mol Cell Biol., 26: 2441-2455, 2006. PMID 16508018
- Truscott M, Denault JB, Goulet B, Leduy L, Salvesen GS, Nepveu A. Carboxyl-terminal proteolytic processing of CUX1 by a caspase enables transcriptional activation in proliferating cells. J Biol Chem., 282: 30216-30226, 2007. PMID 17681953
- Ueda Y, Su Y, Richmond A. CCAAT displacement protein regulates nuclear factor-kappa beta-mediated chemokine transcription in melanoma cells. Melanoma Res., 17: 91-103, 2007. PMID 17496784
- van Gurp MF, Pratap J, Luong M, Javed A, Hoffmann H, Giordano A, Stein JL, Neufeld EJ, Lian JB, Stein GS, van Wijnen AJ. The CCAAT displacement protein/cut homeodomain protein represses osteocalcin gene transcription and forms complexes with the retinoblastoma protein-related protein p107 and cyclin A. Cancer Res., 59: 5980-5988, 1999. PMID 10606245
- Vanden Heuvel GB, Bodmer R, McConnell KR, Nagami GT, Igarashi P. Expression of a cut-related homeobox gene in developing and polycystic mouse kidney. Kidney Int., 50: 453-461, 1996. PMID 8840273


