Methods Type:
Entry Version:
Citation:
Pancreapedia: Exocrine Pancreas Knowledge Base, DOI: 10.3998/panc.2013.10
| Attachment | Size |
|---|---|
| 200.7 KB |
Abstract
Isolating high quality RNA specifically from pancreatic exocrine cells presents a number of challenges for researchers. The bulk of the pancreatic cell mass is composed of acinar cells which synthesize, store and release proteolytic enzymes, RNAses and lipases that contribute to normal digestion. The abundance of these proteins hinders the isolation of high-quality, undegraded RNA from pancreatic exocrine cells and this challenge is further amplified when cells are subjected to Fluorescence Activated Cell Sorting (FACS)-based cell isolation. We present a modified method for pancreatic tissue digestion and FACS sorting of pancreatic cell types. The improvements in RNA quality using this new method are significant, as they enable isolation of high quality RNA from cell types in the pancreas that typically are difficult to sort and extract RNA. This easily adaptable, technically simple method to improve extracted RNA integrity from pancreatic tissues improves cell viability prior to FACS isolation of individual cell populations.
Introduction
Current methods for avoiding the degradation of intact RNA from pancreatic tissues include the use of perfusion techniques to inactivate RNAses using inhibitors (1), the use of commercially available RNAse inhibitors to store tissues prior to RNA extraction (3) and techniques that require rapid freezing prior to tissue processing. Furthermore, a number of methods have been described to isolate ductal cells (6), centroacinar cells (8) and alpha cells from the pancreas (4). Although these protocols yield material that is adequate for RT-PCR and some microarray applications, its use in whole transcriptome analysis by RNA-Sequencing is limited due to the requirement for purified RNA with RNA Integrity Numbers (RIN) greater than eight.
As part of our experimental strategy we sought to explore transcriptome differences among different pancreatic cell types. This experimental strategy required the isolation of RNA from pancreatic ductal and acinar cells. The additional steps involved in the dissociation of whole pancreas and isolation of selected pancreatic cell populations by FACS adds technical challenges that limit both the yield and quality of RNA obtained after processing. We sought to optimize current protocols for pancreatic tissue processing and dissociation prior to FACS isolation of individual cell populations in order to improve the yield of high-quality RNA. This method incorporates the use of a purified form of collagenase and a membrane sealing polaxamer to increase cell viability and prevent the release of pancreatic proteases and RNAses during the preparation and isolation of single cells from whole pancreas. Our group has used this method to successfully isolate high-quality RNA from FACS sorted, highly purified cell populations from the adult and embryonic pancreas suitable for either RNA-Sequencing and microarray applications.
Our protocol combines the use of a purified collagenase enzymes I and II, Liberase DL, in addition to a polaxamer dissolved in tissue culture media to dissociate pancreatic cells. The goal of these modifications was to increase cell viability in our specimens in order to decrease the amount of proteases and RNAses released by injured and dying cells with the ultimate goal of optimizing methods to isolate higher yields of intact RNA for analysis. Current methods rely on the use of crude enzyme preparations of collagenase P, with incubation and washing steps are carried out in salt buffers. We assessed the cell viability and RNA quality of adult murine pancreas preparations using our new method and compared RNA yield and quality to that of the standard collagenase P method (7, 8).
Definitive evidence supporting the benefit of our modified protocol is shown in Figure 1. In this experiment, two samples from the same mouse pancreas were processed with or without the addition of a specific polaxamer, P188 (Note 1). Both samples were run through the machine and cells were isolated. The sorted cells were then analyzed again and the propidium iodide (PI) staining spectrum is shown. Only the cells in a solution of P188 maintained high viability as assessed by PI exclusion. Cells that were not treated with P188 were minimally viable after sorting. We also performed a comparison using a Bioanalyzer of the old method to our new method using Bioanalyzer analysis of RNA integrity and the results are shown in Figure 2. This modified method dramatically increased the quality of the RNA with RIN numbers consistently above eight. Tissue processing using our modified method also resulted in a ten-fold increase in the yield of the RNA, allowing for transcriptome analysis from smaller numbers of isolated cells
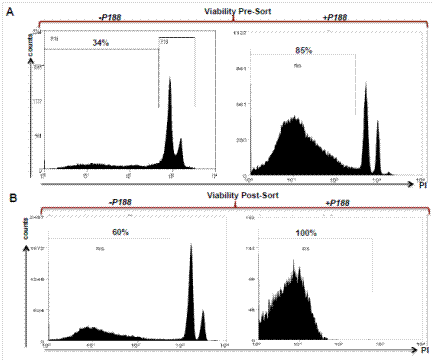
Figure 1. Propidium Iodide exclusion to assess cell viability prior to and after FACS sorting. Cells were prepared and resuspended in solutions with or without P188. Peaks between 103-104 fluorescence intensity determine cell death as measured by loss of membrane integrity with PI staining. The use of P188 significantly increases cell viability post-sort, facilitating the isolation of high quality RNA. Percentages indicate the fraction of viable, PI-excluding cells.
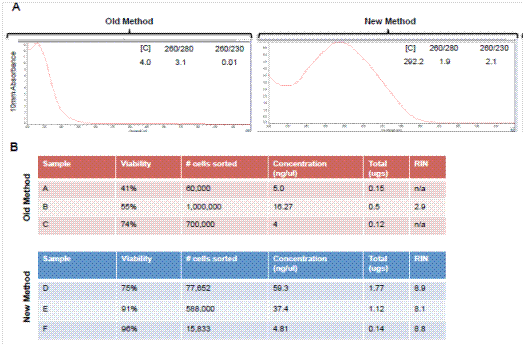
Figure 2. Comparison of yield and quality of RNA from FACS sorted acinar cells from adult murine pancreas. Analyses shown compare traditional methods of tissue preparation and RNA extraction to the newer methods described in this publication. (A) Nanodrop spectra showing spectra of RNA. The new method has the appropriate peak wavelength at 260 nm. (B) Bioanalyzer data showing viability, RNA concentration and RIN numbers from three independent sorting experiments using in the absence (old method) or presence (new method) of P188.
1. Materials
1.1 Reagents
1. MEM without L-glutamine, FBS or antibiotics (Corning)
2. Liberase DL Research Grade (Roche, REF 05 401 160 001), 5mg vial
3. Proteinase inhibitor (Sigma tablets; SigmaFast S8820)
4. Kolliphor® P188 (Sigma; 15759), also known as Pluronic F-68
5. b-mercaptoethanol
6. 1M sterile CaCl2
7. 100 μm mesh (SEFAR; Nitex Nylon)
8. Qiagen RNA lysis buffer (RLT buffer) with fresh b-mercaptoethanol (< 2 years old)
9. Fetal Bovine Serum (FBS)
1.2 Equipment
1. Themo Scientific Nanodrop ND-2000
2. Agilent 2100 Bioanalyzer
3. Orbital Incubator Shaker
4. Eppendorf Centrifuge 5702 R
1.3 Solutions
Prior to day of prep:
1. P188 stock solution (15 mM; 100x stock solution), place in a 37°C water bath to dissolve, filter sterilize and store at 4°C in the dark.
Before euthanizing animal:
2. Make stock solution of enzyme
a. Slowly thaw enzyme (lyophilized form) on ice for 30 min
b. Add 2 ml of MEM
c. Keep on ice, resuspend by pipetting. Do not vortex. This solution can be stored at 4°C for up to a month. Alternatively, aliquot into smaller volumes and store at -20°C. Use or discard an aliquot after thawing-do not re-freeze.
3. Make “A” solution: (Dissociation solution)
a. For each adult whole pancreas digest: make 10 ml of solution
b. 200 mL of stock enzyme solution (Adding more doesn’t increase number of viable cells)
c. 75 ml 1M CaCl2
d. 100 ml P188
e. MEM to 10 ml
f. Place in 37°C water bath
4. Make “C” solution (Cell resuspension solution): Add 2 tablets of proteinase inhibitor to 500 ml bottle of MEM.
5. Make “B” solution (Wash buffer): For 50 ml: 40 ml “C” solution plus 10 ml of FBS.
2. Methods
2.1 Use of a transgenic mouse model for isolation of acinar cells.
Adult murine pancreata were harvested after euthanasia with isofluorane and cervical dislocation in accordance with animal care and use standards. The Mist1:CreERT2 mouse line was utilized to fluorescently label. Adult acinar cells of the murine pancreas (2). This was accomplished by crossing this Cre line to the ROSAmT/MG (mTmG) reporter mouse strain (5). The mTmG mouse strain expresses a membrane-bound form of red fluorescence prior to Cre recombinase and a membrane-bound form of GFP after Cre recombinase. This experimental strategy allows for isolation of acinar cells by FACS by sorting cells that express the highest levels of GFP.
2.2 Pancreas harvest and digestion.
After harvesting, mince each pancreas in 1 mL of dissociation solution A using scissors and then transfer the fragments to a 50 ml conical tube with a final volume of 10 ml of cell dissociation buffer (Note 2). Incubate the samples at 37°C for 30 minutes in an orbital shaker set at 180 rpm. In order to increase cell dissociation, a small sterile glass marble is placed inside the tube and the tube is incubated lying on its side. After 30 min the samples are placed on ice and 10 ml of Wash buffer B is added to each sample. After gently inverting the samples, the digested material is filtered through100 mm nylon mesh and the sample is pelleted at 1,800 rpm for 3 min in a 50ml conical tube at 4°C. The pellet is gently resuspended by pipetting in 5 ml of Wash buffer B followed by another cycle of centrifugation. The pellet is washed with Wash buffer B twice and after the final spin it is resuspended in 1 ml of cell resuspension solution C. The cells are then immediately taken for cell sorting (Notes 3 and 4).
2.3 At flow facility
For cell sorting, it is critically important that you sort only viable cells. ALL samples MUST be analyzed using either PI or a viability dye that doesn’t overlap with the fluors you are trying to use. Exclude non-viable cells from the sort. Any acinar cell that is non-viable or in the process of being non-viable will release its contents and decrease your RNA yield and quality.
It takes 30 minutes to set up parameters in most sorting facilities. Bring samples on ice and bring extra cell resuspension solution in case the samples need to be diluted during the sort. Do not add PBS or other salt containing buffer to the sample and keep it cold. Depending on the total number of cells needed and their % in the sample it can take up to an hour to sort one animal. The amount of time needed to isolate 50,000 cells will depend on the quality of the prep and the number of total cells sorted. When sorting acinar cells, typically 10-30 minutes is needed to isolate 50,000 cells from a dissociated adult pancreas. After sorting ~ 50,000 cells into RLT/b-ME buffer the sample should be removed and mixed before continuing to sort more cells into that tube. This prevents the cells from layering on top of the RLT buffer during the sort run. Isolate RNA as soon as sort is complete. Storing sorted cells either at 4°C or frozen at -80°C leads to decreased RNA yields and low RINs. Typically, 40,000-100,000 cells is enough to yield RNA for subsequent analysis. Finally, sorting a large number of samples is challenging due to the added time for the prep. Limit each sort day to no more than 2 genetically identical samples and no more than 4 animals.
2.4 Cell collection and RNA isolation.
RNA was extracted from pancreatic acinar cells using the Qiagen RNeasy mini kit method immediately after FACS isolation. The cells were kept on ice during and after the sort run. The ratio of cells to RLT buffer was 5x105 cells per 3ml lysis buffer in order to prevent dilution of the RLT buffer with larger sample volumes. After elution from the RNEasy columns the RNA quantity was analyzed using a Nanodrop spectrophotometer (Thermo Scientific) and a bioanalyzer (Agilent).
3. Notes
Note 1: Because the lengths of the polymer blocks can differ the first two digits in the number of the polaxamer x 100 gives the mass of the polypropylene core and the third digit x 10 gives the percentage of polyethylene content.
Note 2 Day of Prep: Be cognizant of the amount of tissue you have. This protocol is optimized for 100 mg of tissue. For 50 mg of tissue, the buffer volumes/enzymes used can by reduced by 50%. Adjust accordingly based on weight and type of tissue: specimens from embryonic tissue will not require as much enzyme or incubation time. Determine empirically.
Note 3: Start 2h before sort time for 2 adult mice if not staining with antibodies. It takes 10-15 min to prep each adult animal so plan appropriately. Add an additional 90 min for the prep if using antibodies
Note 4 If adding antibodies: Resuspend in MEM and add antibodies to cell aliquots as needed. Use: secondary antibody only treated aliquot of cells for FACS control as well as a no antibody control. After staining wash in MEM two times (no PBS or HBSS) and resuspend in cold cell resuspension solution. Transfer to FACS tube and take to facility for analysis and sorting.
Acknowledgements
The authors would like to thank Dr. Hao Zhang for excellent support and recommendations. JMB is supported by the 2011 Pancreatic Action Network-AACR Pathway to Leadership Award and NCI 5F32 CA157044. JA is supported through the 5T32 CA126607 and the immixGroup Foundation Fellowship. SDL is supported by NCI P01 CA134292. SDL is further supported by the Paul K. Neumann Professorship in Pancreatic Cancer at Johns Hopkins University.
4. References
- Griffin M, Abu-El-Haija M, Rokhlina T, and Uc A. Simplified and versatile method for isolation of high-quality RNA from pancreas. Biotechniques 52: 332-334, 2012. PMID: 22578126
- Habbe N, Shi G, Meguid RA, Fendrich V, Esni F, Chen H, Feldmann G, Stoffers DA, Konieczny SF, Leach SD, and Maitra A. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci U S A 105: 18913-18918, 2008. PMID: 19028870
- Itoh T, Sugimoto K, Shimoda M, Chujo D, Takita M, Iwahashi S, Kanak M, Yoshiko T, Naziruddin B, Levy MF, and Matsumoto S. Establishment of a prolonged pancreas preservation model for islet isolation research in mice. Islets 3: 376-380, 2011. PMID: 22045261
- Kohler M, Dare E, Ali MY, Rajasekaran SS, Moede T, Leibiger B, Leibiger IB, Tibell A, Juntti-Berggren L, and Berggren PO. One-step purification of functional human and rat pancreatic alpha cells. Integr Biol (Camb) 4: 209-219, 2012. PMID: 22267247
- Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L. A global double-fluorescent Cre reporter mouse. Genesis 45: 593-605, 2007. PMID:
- Reichert M, Takano S, Heeg S, Bakir B, Botta GP, and Rustgi AK. Isolation, culture and genetic manipulation of mouse pancreatic ductal cells. Nat Protoc 8: 1354-1365, 2013. PMID: 17868096
- Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK, Vonderheide RH, Leach SD, and Stanger BZ. EMT and dissemination precede pancreatic tumor formation. Cell 148: 349-361, 2012. PMID: 22265420
- Rovira M, Scott SG, Liss AS, Jensen J, Thayer SP, and Leach SD. Isolation and characterization of centroacinar/terminal ductal progenitor cells in adult mouse pancreas. Proc Natl Acad Sci U S A 107: 75-80, 2010. PMID: 20018761


